Two New Diphenylketones and a New Xanthone from Talaromyces islandicus EN-501, an Endophytic Fungus Derived from the Marine Red Alga Laurencia okamurai
Abstract
:1. Introduction
2. Results and Discussion
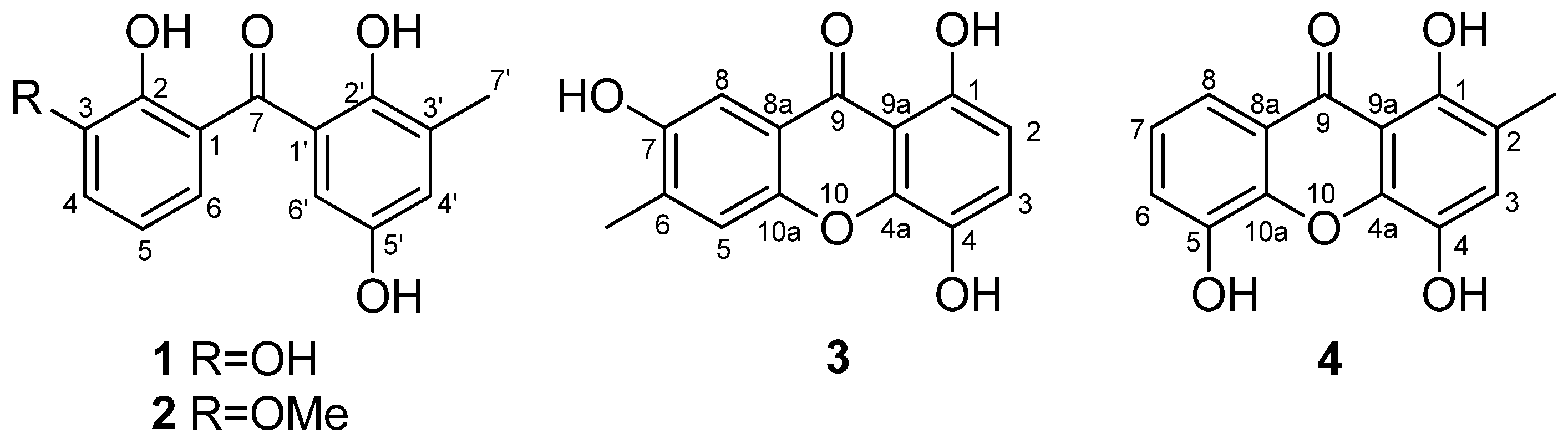
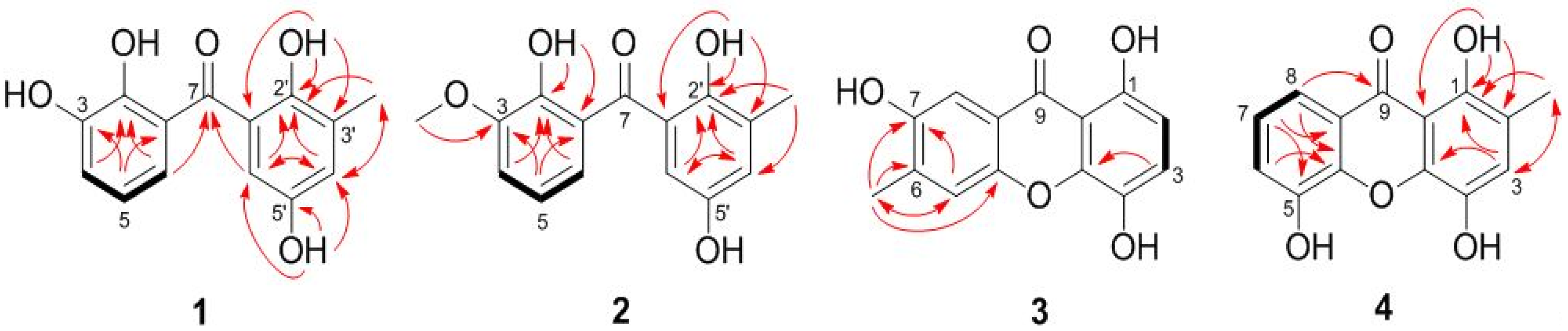
2.1. Structure Elucidation of the New Compounds
2.2. Biological Activities of the Isolated Compounds
3. Experimental Section
3.1. General
3.2. Fungal Material
3.3. Fermentation
3.4. Extraction and Isolation
3.5. X-ray Crystallographic Analysis of Compound 1 [20]
3.6. Antioxidant Assay
3.7. Antimicrobial Assay
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Bugni, T.S.; Ireland, C.M. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004, 21, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar] [CrossRef] [PubMed]
- Du, F.Y.; Zhang, P.; Li, X.M.; Li, C.S.; Cui, C.M.; Wang, B.G. Cyclohexadepsipeptides of the isaridin class from the marine-derived fungus Beauveria felina EN-135. J. Nat. Prod. 2014, 77, 1164–1169. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Li, X.M.; Li, C.S.; Wang, B.G. Diphenyl ether and benzophenone derivatives from the marine mangrove-derived fungus Penicillium sp. MA-37. Phytochem. Lett. 2014, 9, 22–25. [Google Scholar] [CrossRef]
- Meng, L.H.; Zhang, P.; Li, X.M.; Wang, B.G. Penicibrocazines A–E, five new sulfide diketopiperazines from the marine-derived endophytic fungus Penicillium brocae. Mar. Drugs 2015, 13, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Mándi, A.; Li, X.M.; Du, F.Y.; Wang, J.N.; Li, X.; Kurtán, T.; Wang, B.G. Varioxepine A, a 3H-oxepine-containing alkaloid with a new oxa-cage from the marine algal-derived endophytic fungus Paecilomyces variotii. Org. Lett. 2014, 16, 4834–4837. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, X.M.; Wang, J.N.; Li, X.; Wang, B.G. New butenolide derivatives from the marine-derived fungus Paecilomyces variotii with DPPH radical scavenging activity. Phytochem. Lett. 2015, 11, 85–88. [Google Scholar] [CrossRef]
- Zhang, P.; Meng, L.H.; Mándi, A.; Li, X.M.; Kurtán, T.; Wang, B.G. Structure, absolute configuration, and conformational study of resorcylic acid derivatives and related congeners from the fungus Penicillium brocae. RSC Adv. 2015, 5, 39870–39877. [Google Scholar] [CrossRef]
- Xie, X.S.; Fang, X.W.; Huang, R.; Zhang, S.P.; Wei, H.X.; Wu, S.H. A new dimeric anthraquinone from endophytic Talaromyces sp. YE3016. Nat. Prod. Res. 2016, 30, 1706–1711. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.C.; Yang, Z.D.; Zhou, S.Y.; Yu, H.T.; Zhang, F.; Yao, X.J. Two new compounds, deacetylisowortmins A ans B, isolated from an endophytic fungus, Talaromyces wortmannii LGT-4. Nat. Prod. Res. 2016, 30, 1623–1627. [Google Scholar] [CrossRef] [PubMed]
- Zhi, K.K.; Yang, Z.D.; Zhou, S.Y.; Yao, X.J.; Li, S.; Zhang, F. A new furanosteroid from Talaromyces sp. lgt-4, a fungal endophyte isolated from Tripterygium wilfordii. Nat. Prod. Res. 2016, 30, 2137–2141. [Google Scholar] [PubMed]
- Rudman, P.; Gay, F.J. The Causes of Natural Durability in Timber X. Holzforschung 1963, 17, 21–25. [Google Scholar] [CrossRef]
- Fouotsa, H.; Lannang, A.M.; Dzoyem, J.P.; Tatsimo, S.J.N.; Neumann, B.; Mbazoa, C.D.; Razakarivony, A.A.; Nkengfack, A.E.; Eloff, J.N.; Sewald, N. Antibacterial and Antioxidant Xanthones and Benzophenone from Garcinia smeathmannii. Planta Med. 2015, 81, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Iinuma, M.; Tosa, H.; Ito, T.; Tanaka, T.; Madulid, D.A. Two xanthones from roots of Cratoxylum formosanum. Phytochemistry 1996, 42, 1195–1198. [Google Scholar] [CrossRef]
- Le Pogam, P.; Boustie, J. Xanthones of lichen source: A 2016 update. Molecules 2016, 21, 294. [Google Scholar] [CrossRef] [PubMed]
- Wezeman, T.; Bräse, S.; Masters, K.S. Xanthone dimers: A compound family which is both common and privileged. Nat. Prod. Rep. 2015, 32, 6–28. [Google Scholar] [CrossRef] [PubMed]
- Dieu, A.; Millot, M.; Champavier, Y.; Mambu, L.; Chaleix, V.; Sol, V.; Gloaguen, V. Uncommon chlorinated xanthone and other antibacterial compounds from the lichen Cladonia incrassata. Planta Med. 2014, 80, 931–935. [Google Scholar] [CrossRef] [PubMed]
- Rezanka, T.; Sigler, K. Hirtusneanoside, an unsymmetrical dimeric tetrahydroxanthone from the lichen Usnea hirta. J. Nat. Prod. 2007, 70, 1487–1491. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, X.M.; Teuscher, F.; Li, D.L.; Diesel, A.; Ebel, R.; Proksch, P.; Wang, B.G. Chaetopyranin, a benzaldehyde derivative, and other related metabolites from Chaetomium globosum, an endophytic fungus derived from the marine red alga Polysiphonia urceolata. J. Nat. Prod. 2006, 69, 1622–1625. [Google Scholar] [CrossRef] [PubMed]
- Crystallographic data of compound 1 have been deposited in the Cambridge Crystallographic Data Centre as CCDC 1465271. The data can be obtained free of charge via http://www.ccdc.cam.ac.uk/data_request/cif, by emailing [email protected], or by contacting the CCDC, 12 Union Road, Cambridge CB21EZ, UK; Fax: +44-1223-336-033.
- Sheldrick, G.M. SADABS, Software for Empirical Absorption Correction; University of Göttingen: Göttingen, Germany, 1996. [Google Scholar]
- Sheldrick, G.M. SHELXTL, Structure Determination Software Programs; Bruker Analytical X-ray System Inc.: Madison, WI, USA, 1997. [Google Scholar]
- Sheldrick, G.M. SHELXL-97 and SHELXS-97, Program for X-ray Crystal Structure Solution and Refinement; University of Göttingen: Göttingen, Germany, 1997. [Google Scholar]
- Pierce, C.G.; Uppuluri, P.; Tristan, A.R.; Wormley, F.L., Jr.; Mowat, E.; Ramage, G.; Lopez-Ribot, J.L. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 2008, 3, 1494–1500. [Google Scholar] [CrossRef] [PubMed]

| Position | 1 (Measured in DMSO-d6) | 2 (Measured in CDCl3) | ||
|---|---|---|---|---|
| ΔC | ΔH | ΔC | ΔH | |
| 1 | 126.1, C | 122.1, C | ||
| 2 | 143.8, C | 148.5, C | ||
| 3 | 145.8, C | 148.2, C | ||
| 4 | 117.7, CH | 6.98, d (7.7) | 123.1, CH | 7.11, d (7.0) |
| 5 | 119.2, CH | 6.77, t (7.7) | 118.9, CH | 6.89, t (7.2) |
| 6 | 119.0, CH | 6.71, d (7.6) | 115.2, CH | 7.04, d (7.0) |
| 7 | 202.5, C | 201.7, C | ||
| 1′ | 120.2, C | 119.0, C | ||
| 2′ | 152.3, C | 155.0, C | ||
| 3′ | 127.1, C | 129.0, C | ||
| 4′ | 125.3, CH | 6.91, br s | 125.6, CH | 6.94, br s |
| 5′ | 148.7, C | 146.5, C | ||
| 6′ | 114.8, CH | 6.62, br s | 115.5, CH | 6.84, br s |
| 7′ | 15.4, CH3 | 2.18, s | 15.8, CH3 | 2.27, s |
| 2-OH | 9.00, s | |||
| 3-OH/OMe | 9.47, s | 56.3, CH3 | 3.93, s | |
| 2′-OH | 11.42, s | 10.95, s | ||
| 5′-OH | 9.02, s | 4.57, s | ||
| Position | 3 (Measured in DMSO-d6) | 4 (Measured in DMSO-d6) | ||
|---|---|---|---|---|
| ΔC | ΔH | ΔC | ΔH | |
| 1 | 152.0, C | 149.7, C | ||
| 2 | 110.6, CH | 7.42, d (7.2) | 117.6, C | |
| 3 | 124.5, CH | 7.24, d (7.2) | 124.6, CH | 7.21, s |
| 4 | 137.9, C | 137.0, C | ||
| 4a | 146.2, C | 141.0, C | ||
| 5 | 123.8, CH | 7.20, s | 147.6, C | |
| 6 | 117.1, C | 120.8, CH | 7.32, d (7.6) | |
| 7 | 149.1, C | 124.3, CH | 7.27, t (7.8) | |
| 8 | 120.9, CH | 7.26, s | 113.8, CH | 7.54, d (7.7) |
| 8a | 120.5, C | 120.6, C | ||
| 9 | 182.8, C | 182.3, C | ||
| 9a | 107.7, C | 107.8, C | ||
| 10a | 141.2, C | 145.1, C | ||
| 2-CH3 | 14.4, CH3 | 2.17, s | ||
| 6-CH3 | 14.5, CH3 | 2.19, s | ||
| 1-OH | 12.04, s | 12.06, s | ||
| Samples | 1 | 2 | 3 | 4 | BHT a | Ascorbic Acid |
|---|---|---|---|---|---|---|
| DPPH | 1.26 | 1.33 | 6.92 | 1.23 | 16.27 | |
| ABTS | 0.69 | 0.58 | 2.35 | 1.27 | 3.01 |
| Samples | 1 | 2 | 3 | 4 | Chloramphenicol |
|---|---|---|---|---|---|
| EC | 4 | >64 | 32 | 4 | 1 |
| PA | 4 | >64 | 32 | 4 | 4 |
| SA | 8 | >64 | >64 | 8 | 2 |
| VA | 4 | >64 | 32 | 4 | 0.5 |
| VH | 8 | >64 | 32 | 8 | 2 |
| VP | 4 | >64 | 32 | 4 | 2 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.-L.; Li, X.-M.; Liu, H.; Meng, L.-H.; Wang, B.-G. Two New Diphenylketones and a New Xanthone from Talaromyces islandicus EN-501, an Endophytic Fungus Derived from the Marine Red Alga Laurencia okamurai. Mar. Drugs 2016, 14, 223. https://doi.org/10.3390/md14120223
Li H-L, Li X-M, Liu H, Meng L-H, Wang B-G. Two New Diphenylketones and a New Xanthone from Talaromyces islandicus EN-501, an Endophytic Fungus Derived from the Marine Red Alga Laurencia okamurai. Marine Drugs. 2016; 14(12):223. https://doi.org/10.3390/md14120223
Chicago/Turabian StyleLi, Hong-Lei, Xiao-Ming Li, Hui Liu, Ling-Hong Meng, and Bin-Gui Wang. 2016. "Two New Diphenylketones and a New Xanthone from Talaromyces islandicus EN-501, an Endophytic Fungus Derived from the Marine Red Alga Laurencia okamurai" Marine Drugs 14, no. 12: 223. https://doi.org/10.3390/md14120223






