Deep Subseafloor Fungi as an Untapped Reservoir of Amphipathic Antimicrobial Compounds
Abstract
:1. Introduction
2. Results and Discussion
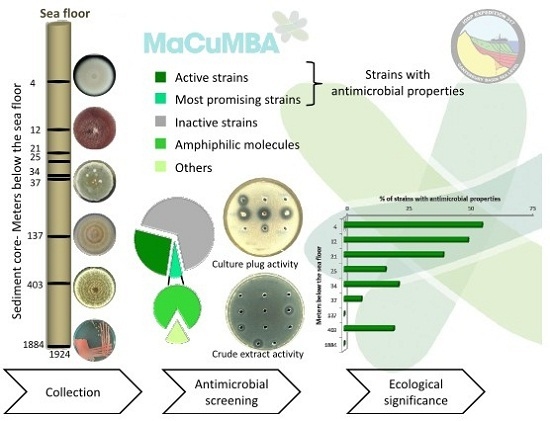
2.1. Antimicrobial Screening
- Anti Gram-positive fungi, as the most important group clustering 15 strains (53% of the bioactive strains). Isolates were identified as belonging to Aspergillus fumigatus 48X3-P3-P1(2), Aspergillus terreus 1H3-S0-P1(1), Eurotium herbariorum CB_33, Fusarium oxysporum (1H3-P0-P1(1), 4H1-P0-P1(1) and 4H1-P3-P3), Penicillium bialowiezense (CB_5, CB_7 and CB_8), Penicillium chrysogenum (2H5-M3-P2-(3), CB_11, CB_17 and CB_24), Penicillium sp. CB_16 (1 strain), and Oidiodendron griseum CB_36 species. Marine species of Penicillium, Aspergillus and Fusarium genera are well-known as producers of a wide array of bioactive metabolites, i.e., polyketides, alkaloids and peptides, with a broad spectrum of biological activities [12,26,27,28]. Several marine-derived Eurotium species have been investigated for antimicrobial activity with a special focus on Eurotium cristatum. An algae-associated E. cristatum strain was assessed as producer of the tardioxopiperazine A antibiotic inhibiting Staphylococcus aureus [29] contrary to one sponge-associated E. cristatum strain [30]. Interestingly, and to the best of knowledge, no antimicrobial activity was ever detected from marine-derived Oidiodendron strains while, some terrestrial strains, as Oidiodendron truncatum or Oidiodendron fuscum, are able to synthesize antibacterial polyketide (fuscin) [31] and terpenoid (clerocidin) [32].
- Anti-fungal strains gathering: Penicillium bialowiezense (CB_4, CB_5, CB_6, CB_7, CB_8, CB_9 and CB_10), Sarocladium sp (2H5-S0-P7(2), 3H5-P3-P6(1) and 5H3-M3-P2), Fusarium oxysporum (4H1-P0-P1(1) and 4H1-P3-P3) and Paecilomyces sp. 1H3-M3-P1(2) species. Marine species of Penicillium and Fusarium genera have already been described as producers of antifungal terpenoids like penicisteroids and polyketides as fusarielin E, respectively [33,34,35,36,37,38]. Marine-derived Paecilomyces species, isolated from mangrove habitats, have been shown to produce antifungal polyketides [39,40]. Sarocladium kiliense and Sarocladium oryzae are known to produce antifungal terpenoid and polyketide against phytopathogenic fungi [41] and C. albicans [42,43]. A marine endophytic Sarocladium sp. was described for its ability to inhibit bacterial quorum sensing [44]. Here, we report an original ability of a deep subseafloor Sarocladium isolate to biosynthesize bioactive compounds against A. flavus.
- Five deep subseafloor fungal isolates had a weak antibacterial activity against Gram-negative bacteria (partial inhibition). Those isolates were identified as Acremonium sp. 5H1-M3-P8(1), Aspergillus terreus 1H3-S0-P1(1), Fusarium oxysporum 5H1-S0-P1(3), Sistotrema brinkmanii 5H1-P0-P5(1)bis and an uncultured Agaricomycete CB_23. If species of Aspergillus, Fusarium and Acremonium genera have already been described to produce an array of anti-Gram-negative compounds [45,46,47,48,49,50], to the best of knowledge we here described an original antimicrobial activity from a Sistotrema brinkmannii species.
2.2. Ecological Significance
2.3. Antimicrobial Compounds Production and Extraction
3. Materials and Methods
3.1. Strains Collection
3.2. Cultivation Methods
3.3. Antimicrobial Screening
3.4. Extraction of Amphipathic Bioactive Compounds
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Frieden, T. Antibiotic Resistance Threats in the United States, 2013. Available online: http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (accessed on 20 November 2015).
- Monnet, D.L.; Albiger, B. European Centre for Disease Prevention and Control. Available online: http://ecdc.europa.eu/en/activities/diseaseprogrammes/ARHAI/Pages/about_programme.aspx (accessed on 10 November 2015).
- Brooks, B.D.; Brooks, A.E. Therapeutic strategies to combat antibiotic resistance. Adv. Drug Deliv. Rev. 2014, 78, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 Years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. (Tokyo) 2012, 65, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Endo, A.; Kuroda, M.; Tsujita, Y. ML-236A, ML-236B, and ML-236C, new inhibitors of cholesterogensis produced by Penicillium citrinum. J. Antibiot. (Tokyo) 1976, 29, 1346–1348. [Google Scholar] [CrossRef] [PubMed]
- Fliri, H.; Baumann, G.; Enz, A.; Kallen, J.; Luyten, M.; Mikol, V.; Movva, R.; Quesniaux, V.; Schreier, M.; Walkinshaw, M.; et al. Cyclosporins. Structure-activity relationships. Ann. N. Y. Acad. Sci. 1993, 696, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Debbab, A.; Aly, A.H.; Lin, W.H.; Proksch, P. Bioactive compounds from marine bacteria and fungi: Marine bioactive compounds. Microb. Biotechnol. 2010, 3, 544–563. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar] [CrossRef] [PubMed]
- Ciobanu, M.-C.; Burgaud, G.; Dufresne, A.; Breuker, A.; Rédou, V.; Ben Maamar, S.; Gaboyer, F.; Vandenabeele-Trambouze, O.; Lipp, J.S.; Schippers, A.; et al. Microorganisms persist at record depths in the subseafloor of the Canterbury Basin. ISME J. 2014, 8, 1370–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hay, M.E. Marine chemical ecology: Chemical signals and cues structure marine populations, communities, and ecosystems. Annu. Rev. Mar. Sci. 2009, 1, 193–212. [Google Scholar] [CrossRef] [PubMed]
- Rédou, V.; Navarri, M.; Meslet-Cladière, L.; Barbier, G.; Burgaud, G. Species richness and adaptation of marine fungi from deep-subseafloor sediments. Appl. Environ. Microbiol. 2015, 81, 3571–3583. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, P.; Keller, N.P. Strategies for mining fungal natural products. J. Ind. Microbiol. Biotechnol. 2014, 41, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Golubev, W.I. Antagonistic interactions among yeasts. In Biodiversity and Ecophysiology of Yeasts; Péter, G., Rosa, C., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2006; pp. 197–219. [Google Scholar]
- Vaz, A.B.M.; Mota, R.C.; Bomfim, M.R.Q.; Vieira, M.L.A.; Zani, C.L.; Rosa, C.A.; Rosa, L.H. Antimicrobial activity of endophytic fungi associated with Orchidaceae in Brazil. Can. J. Microbiol. 2009, 55, 1381–1391. [Google Scholar] [CrossRef] [PubMed]
- Coda, R.; Rizzello, C.G.; Di Cagno, R.; Trani, A.; Cardinali, G.; Gobbetti, M. Antifungal activity of Meyerozyma guilliermondii: Identification of active compounds synthesized during dough fermentation and their effect on long-term storage of wheat bread. Food Microbiol. 2013, 33, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Joel, E.L.; Valentin Bhimba, B. Evaluation of secondary metabolites from mangrove associated fungi Meyerozyma guilliermondii. Alex. J. Med. 2013, 49, 189–194. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Bao, J.; Wang, G.-H.; He, F.; Xu, X.-Y.; Qi, S.-H. Diversity and antimicrobial activity of culturable fungi isolated from six species of the South China Sea gorgonians. Microb. Ecol. 2012, 64, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Y.; Zhang, Y.; Xu, X.-Y.; Qi, S.-H. Diverse deep-sea fungi from the South China Sea and their antimicrobial activity. Curr. Microbiol. 2013, 67, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Henríquez, M.; Vergara, K.; Norambuena, J.; Beiza, A.; Maza, F.; Ubilla, P.; Araya, I.; Chávez, R.; San-Martín, A.; Darias, J.; et al. Diversity of cultivable fungi associated with Antarctic marine sponges and screening for their antimicrobial, antitumoral and antioxidant potential. World J. Microbiol. Biotechnol. 2014, 30, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.-Y.; Yang, K.-L.; Li, J.; Wang, C.-Y.; Shao, C.-L. Phylogenetic diversity and antibacterial activity of culturable fungi derived from the Zoanthid Palythoa haddoni in the South China Sea. Mar. Biotechnol. 2015, 17, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, L.A.; Tanaka, N.; Kalinovskaya, N.I.; Mikhailov, V.V. Antimicrobial potential of deep surface sediment associated bacteria from the Sea of Japan. World J. Microbiol. Biotechnol. 2013, 29, 1169–1177. [Google Scholar] [CrossRef] [PubMed]
- Schueffler, A.; Anke, T. Fungal natural products in research and development. Nat. Prod. Rep. 2014, 31, 1425–1448. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Meng, W.; Cao, C.; Wang, J.; Shan, W.; Wang, Q. Antibacterial and antifungal compounds from marine fungi. Mar. Drugs 2015, 13, 3479–3513. [Google Scholar] [CrossRef] [PubMed]
- Du, F.-Y.; Li, X.-M.; Li, C.-S.; Shang, Z.; Wang, B.-G. Cristatumins A–D, new indole alkaloids from the marine-derived endophytic fungus Eurotium cristatum EN-220. Bioorg. Med. Chem. Lett. 2012, 22, 4650–4653. [Google Scholar] [CrossRef] [PubMed]
- Gomes, N.M.; Dethoup, T.; Singburaudom, N.; Gales, L.; Silva, A.M.S.; Kijjoa, A. Eurocristatine, a new diketopiperazine dimer from the marine sponge-associated fungus Eurotium cristatum. Phytochem. Lett. 2012, 5, 717–720. [Google Scholar] [CrossRef]
- Michael, S.E. Studies in the biochemistry of micro-organisms. 79. Fuscin, a metabolic product of Oidiodendron fuscum Robak. Part 1. Preparation, properties and antibacterial activity. Biochem. J. 1948, 43, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Andersen, N.R.; Lorck, H.O.B.; Rasmussen, P.R. Fermentation, isolation and characterization of antibiotic PR-1350. J. Antibiot. (Tokyo) 1983, 36, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.; Zhao, L.L.; Hu, C.Q.; Zhang, H.P. Fusarielin E, a new antifungal antibiotic from Fusarium sp. Chin. Chem. Lett. 2007, 18, 954–956. [Google Scholar] [CrossRef]
- Shao, C.; Wang, C.; Wei, M.; Gu, Y.; Xia, X.; She, Z.; Lin, Y. Structure elucidation of two new xanthone derivatives from the marine fungus Penicillium sp. (ZZF 32#) from the South China Sea. Magn. Reson. Chem. 2008, 46, 1066–1069. [Google Scholar] [PubMed]
- Trisuwan, K.; Rukachaisirikul, V.; Sukpondma, Y.; Phongpaichit, S.; Preedanon, S.; Sakayaroj, J. Lactone derivatives from the marine-derived fungus Penicillium sp. PSU-F44. Chem. Pharm. Bull. (Tokyo) 2009, 57, 1100–1102. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.-S.; Li, X.-M.; Li, C.-S.; Proksch, P.; Wang, B.-G. Penicisteroids A and B, antifungal and cytotoxic polyoxygenated steroids from the marine alga-derived endophytic fungus Penicillium chrysogenum QEN-24S. Bioorg. Med. Chem. Lett. 2011, 21, 2894–2897. [Google Scholar] [CrossRef] [PubMed]
- Park, M.S.; Fong, J.J.; Oh, S.-Y.; Kwon, K.K.; Sohn, J.H.; Lim, Y.W. Marine-derived Penicillium in Korea: Diversity, enzyme activity, and antifungal properties. Antonie van Leeuwenhoek 2014, 106, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Hussain, H.; Drogies, K.-H.; Al-Harrasi, A.; Hassan, Z.; Shah, A.; Rana, U.A.; Green, I.R.; Draeger, S.; Schulz, B.; Krohn, K. Antimicrobial constituents from endophytic fungus Fusarium sp. Asian Pac. J. Trop. Dis. 2015, 5, 186–189. [Google Scholar] [CrossRef]
- Wen, L.; Lin, Y.-C.; She, Z.-G.; Du, D.-S.; Chan, W.-L.; Zheng, Z.-H. Paeciloxanthone, a new cytotoxic xanthone from the marine mangrove fungus Paecilomyces sp. (Tree1-7). J. Asian Nat. Prod. Res. 2008, 10, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Mosadeghzad, Z.; Zuriati, Z.; Asmat, A.; Gires, U.; Wickneswari, R.; Pittayakhajonwut, P.; Farahani, G.H.N. Chemical components and bioactivity of the marine-derived fungus Paecilomyces sp. Collected from Tinggi Island, Malaysia. Chem. Nat. Compd. 2013, 49, 621–625. [Google Scholar] [CrossRef]
- Lou, J.; Fu, L.; Luo, R.; Wang, X.; Luo, H.; Zhou, L. Endophytic fungi from medicinal herb Salvia miltiorrhiza Bunge and their antimicrobial activity. Afr. J. Microbiol. Res. 2013, 7, 5343–5349. [Google Scholar]
- Sakthivel, N.; Gnanamanickam, S. Isolation of and assay for cerulenin produced by sheath rot pathogen, Sarocladium oryzae (Saw.) Gams. Curr. Sci. 1986, 55, 988–989. [Google Scholar]
- Tschen, J.S.-M.; Chen, L.-L.; Hsieh, S.-T.; Wu, T. Isolation and phytotoxic effects of helvolic acid from plant pathogenic fungus Sarocladium oryzae. Bot. Bull. Acad. Sin. 1997, 38, 251–256. [Google Scholar]
- Martín-Rodríguez, A.J.; Reyes, F.; Martín, J.; Pérez-Yépez, J.; León-Barrios, M.; Couttolenc, A.; Espinoza, C.; Trigos, Á.; Martín, V.S.; Norte, M.; et al. Inhibition of bacterial quorum sensing by extracts from aquatic fungi: First report from marine endophytes. Mar. Drugs 2014, 12, 5503–5526. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, J.; Liu, P.; Wang, W.; Zhu, W. Three new compounds from Aspergillus terreus PT06-2 grown in a high salt medium. Mar. Drugs 2011, 9, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Julianti, E.; Oh, H.; Jang, K.H.; Lee, J.K.; Lee, S.K.; Oh, D.-C.; Oh, K.-B.; Shin, J. Acremostrictin, a highly oxygenated metabolite from the marine fungus Acremonium strictum. J. Nat. Prod. 2011, 74, 2592–2594. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xu, Y.; Shao, C.-L.; Yang, R.-Y.; Zheng, C.-J.; Chen, Y.-Y.; Fu, X.-M.; Qian, P.-Y.; She, Z.-G.; de Voogd, N.J.; et al. Antibacterial bisabolane-type sesquiterpenoids from the sponge-derived fungus Aspergillus sp. Mar. Drugs 2012, 10, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Y.; Xu, X.-Y.; Peng, J.; Ma, C.-F.; Nong, X.-H.; Bao, J.; Zhang, G.-Z.; Qi, S.-H. Antifouling potentials of eight deep-sea-derived fungi from the South China Sea. J. Ind. Microbiol. Biotechnol. 2014, 41, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, R.; Mathew, A.; Purohit, H.J. Characterization of antibacterial activity of bikaverin from Fusarium sp. HKF15. J. Biosci. Bioeng. 2014, 117, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Shiono, Y.; Ogata, K.; Koseki, T.; Murayama, T.; Funakoshi, T. A cleistanthane diterpene from a marine-derived fusarium species under submerged fermentation. Z. Für Naturforschung B 2014, 65, 753–756. [Google Scholar] [CrossRef]
- Parkes, R.J.; Cragg, B.A.; Wellsbury, P. Recent studies on bacterial populations and processes in subseafloor sediments: A review. Hydrogeol. J. 2000, 8, 11–28. [Google Scholar] [CrossRef]
- D’Hondt, S.; Spivack, A.J.; Pockalny, R.; Ferdelman, T.G.; Fischer, J.P.; Kallmeyer, J.; Abrams, L.J.; Smith, D.C.; Graham, D.; Hasiuk, F.; et al. Subseafloor sedimentary life in the south Pacific Gyre. Proc. Natl. Acad. Sci. 2009, 106, 11651–11656. [Google Scholar] [CrossRef] [PubMed]
- Kallmeyer, J.; Pockalny, R.; Adhikari, R.R.; Smith, D.C.; D’Hondt, S. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. 2012, 109, 16213–16216. [Google Scholar] [CrossRef] [PubMed]
- Burkepile, D.E.; Parker, J.D.; Woodson, C.B.; Mills, H.J.; Kubanek, J.; Sobecky, P.A.; Hay, M.E. Chemically mediated competition between microbes and animals: Microbes as consumers in food webs. Ecology 2006, 87, 2821–2831. [Google Scholar] [CrossRef]
- Taylor, M.W.; Hill, R.T.; Piel, J.; Thacker, R.W.; Hentschel, U. Soaking it up: The complex lives of marine sponges and their microbial associates. ISME J. 2007, 1, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Strom, S.L. Microbial ecology of ocean biogeochemistry: A community perspective. Science 2008, 320, 1043–1045. [Google Scholar] [CrossRef] [PubMed]
- Bigelis, R.; He, H.; Yang, H.Y.; Chang, L.-P.; Greenstein, M. Production of fungal antibiotics using polymeric solid supports in solid-state and liquid fermentation. J. Ind. Microbiol. Biotechnol. 2006, 33, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Hölker, U.; Lenz, J. Solid-state fermentation—Are there any biotechnological advantages? Curr. Opin. Microbiol. 2005, 8, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Barrios-González, J.; Baños, J.G.; Covarrubias, A.A.; Garay-Arroyo, A. Lovastatin biosynthetic genes of Aspergillus terreus are expressed differentially in solid-state and in liquid submerged fermentation. Appl. Microbiol. Biotechnol. 2008, 79, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Souchothèque. Available online: http://www.univ-brest.fr/souchotheque (accessed on 10 November 2015).
- Defer, D.; Desriac, F.; Henry, J.; Bourgougnon, N.; Baudy-Floc, H.M.; Brillet, B.; Le Chevalier, P.; Fleury, Y. Antimicrobial peptides in oyster hemolymph: The bacterial connection. Fish Shellfish Immunol. 2013, 34, 1439–1447. [Google Scholar] [CrossRef] [PubMed] [Green Version]



 : size of the inhibition zone 2, 3 and 5 mm. ✓ Presence, or ✗ absence of genes coding PKS I, PKS III, NRPS, PKS-NRPS and TPS [15]. F10-90: C18-SPE fraction eluted with acetonitrile 90%. OP: Organic phase. In bracket the concentration (mg/mL) of the crude extract. Na: Not assayed
: size of the inhibition zone 2, 3 and 5 mm. ✓ Presence, or ✗ absence of genes coding PKS I, PKS III, NRPS, PKS-NRPS and TPS [15]. F10-90: C18-SPE fraction eluted with acetonitrile 90%. OP: Organic phase. In bracket the concentration (mg/mL) of the crude extract. Na: Not assayed
 : size of the inhibition zone 2, 3 and 5 mm. ✓ Presence, or ✗ absence of genes coding PKS I, PKS III, NRPS, PKS-NRPS and TPS [15]. F10-90: C18-SPE fraction eluted with acetonitrile 90%. OP: Organic phase. In bracket the concentration (mg/mL) of the crude extract. Na: Not assayed
: size of the inhibition zone 2, 3 and 5 mm. ✓ Presence, or ✗ absence of genes coding PKS I, PKS III, NRPS, PKS-NRPS and TPS [15]. F10-90: C18-SPE fraction eluted with acetonitrile 90%. OP: Organic phase. In bracket the concentration (mg/mL) of the crude extract. Na: Not assayed
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navarri, M.; Jégou, C.; Meslet-Cladière, L.; Brillet, B.; Barbier, G.; Burgaud, G.; Fleury, Y. Deep Subseafloor Fungi as an Untapped Reservoir of Amphipathic Antimicrobial Compounds. Mar. Drugs 2016, 14, 50. https://doi.org/10.3390/md14030050
Navarri M, Jégou C, Meslet-Cladière L, Brillet B, Barbier G, Burgaud G, Fleury Y. Deep Subseafloor Fungi as an Untapped Reservoir of Amphipathic Antimicrobial Compounds. Marine Drugs. 2016; 14(3):50. https://doi.org/10.3390/md14030050
Chicago/Turabian StyleNavarri, Marion, Camille Jégou, Laurence Meslet-Cladière, Benjamin Brillet, Georges Barbier, Gaëtan Burgaud, and Yannick Fleury. 2016. "Deep Subseafloor Fungi as an Untapped Reservoir of Amphipathic Antimicrobial Compounds" Marine Drugs 14, no. 3: 50. https://doi.org/10.3390/md14030050