Molecular Characterization of Voltage-Gated Sodium Channels and Their Relations with Paralytic Shellfish Toxin Bioaccumulation in the Pacific Oyster Crassostrea gigas
Abstract
:1. Introduction
2. Results
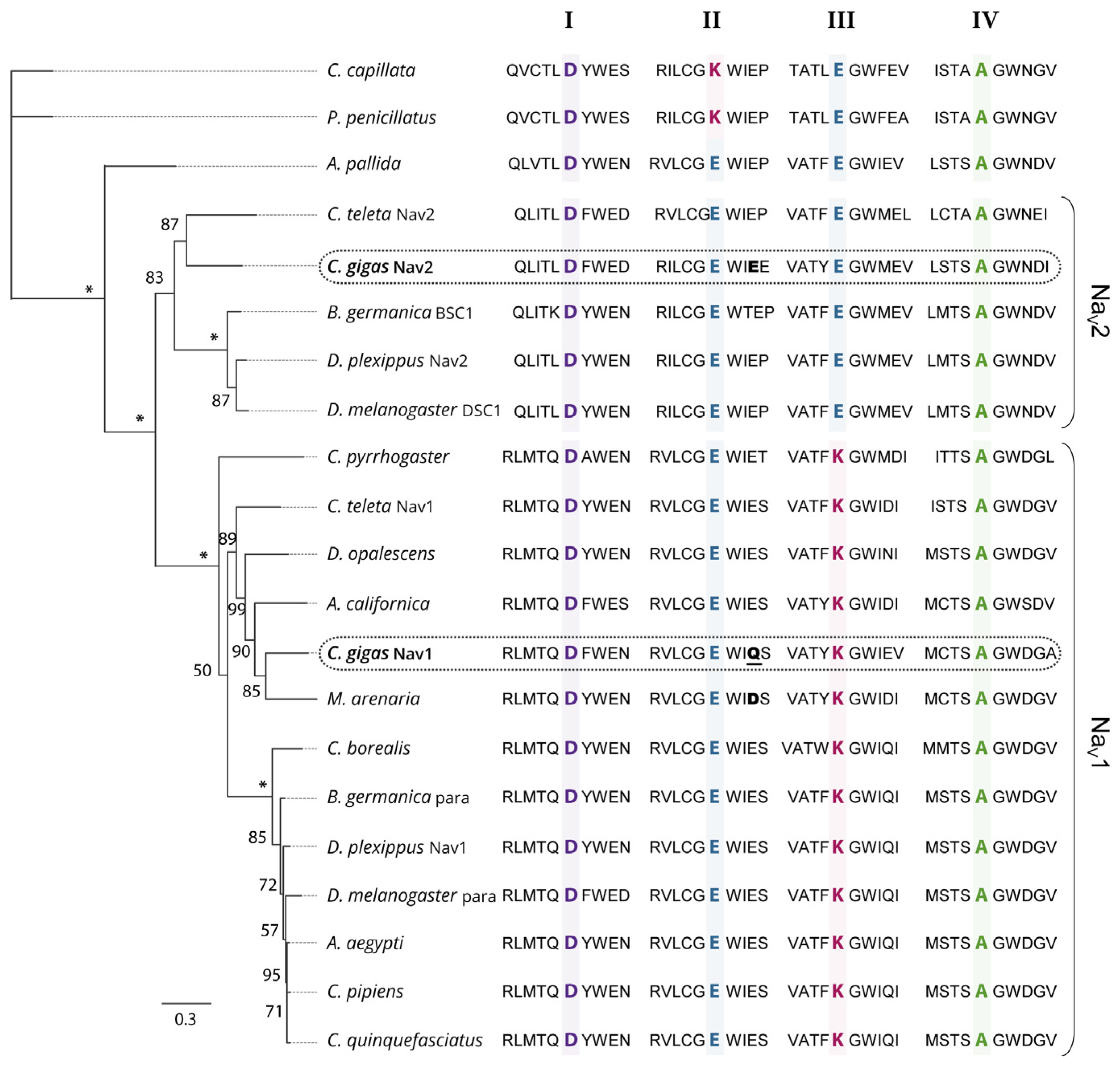
2.1. Phylogenetic Analysis of Nav Channels in Crassostrea gigas
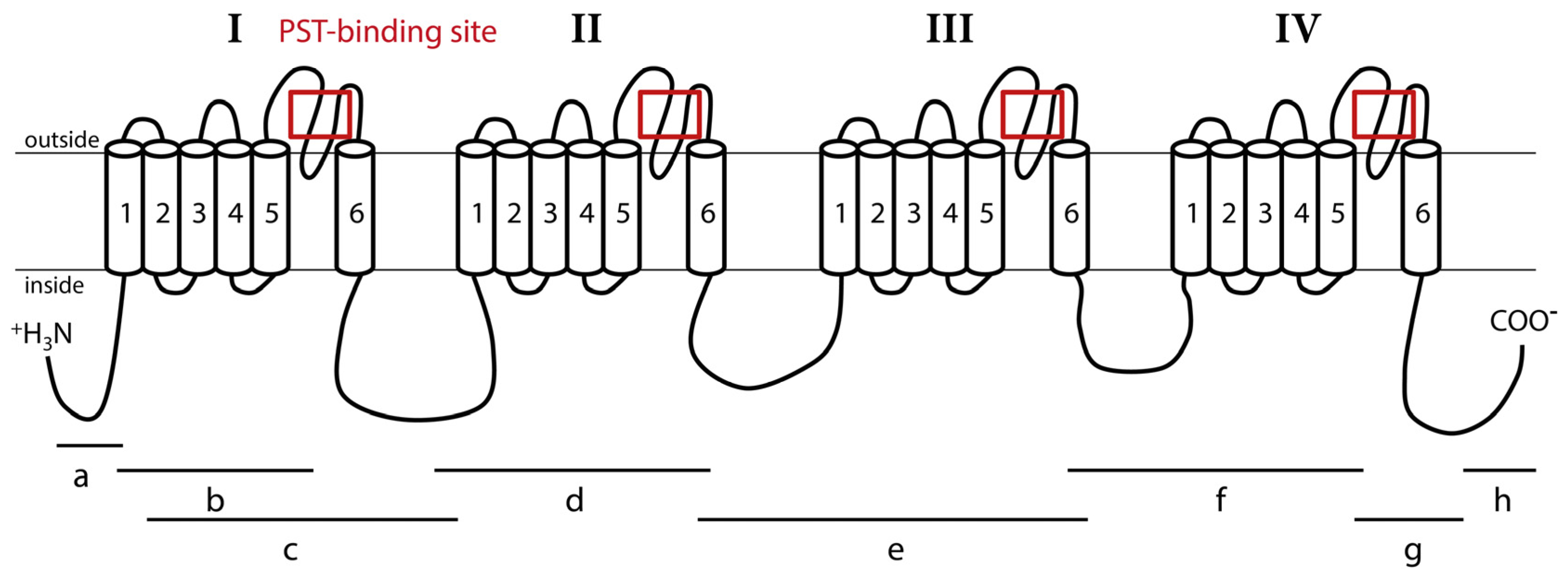
2.2. Structure of CgNav1 α Subunit
2.3. CgNav1 DNA Polymorphism
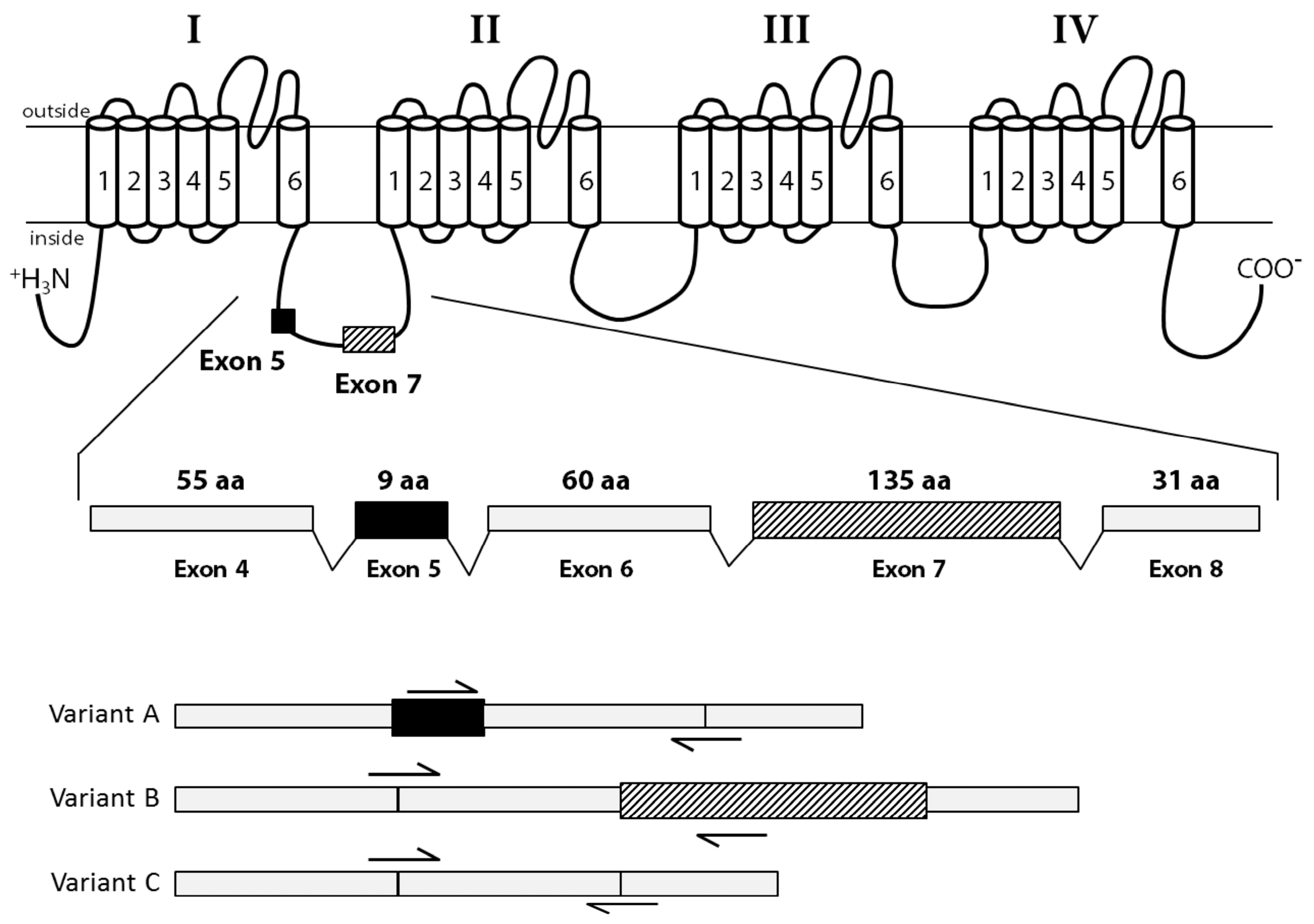
2.4. Identification of CgNav1 Splice Variants
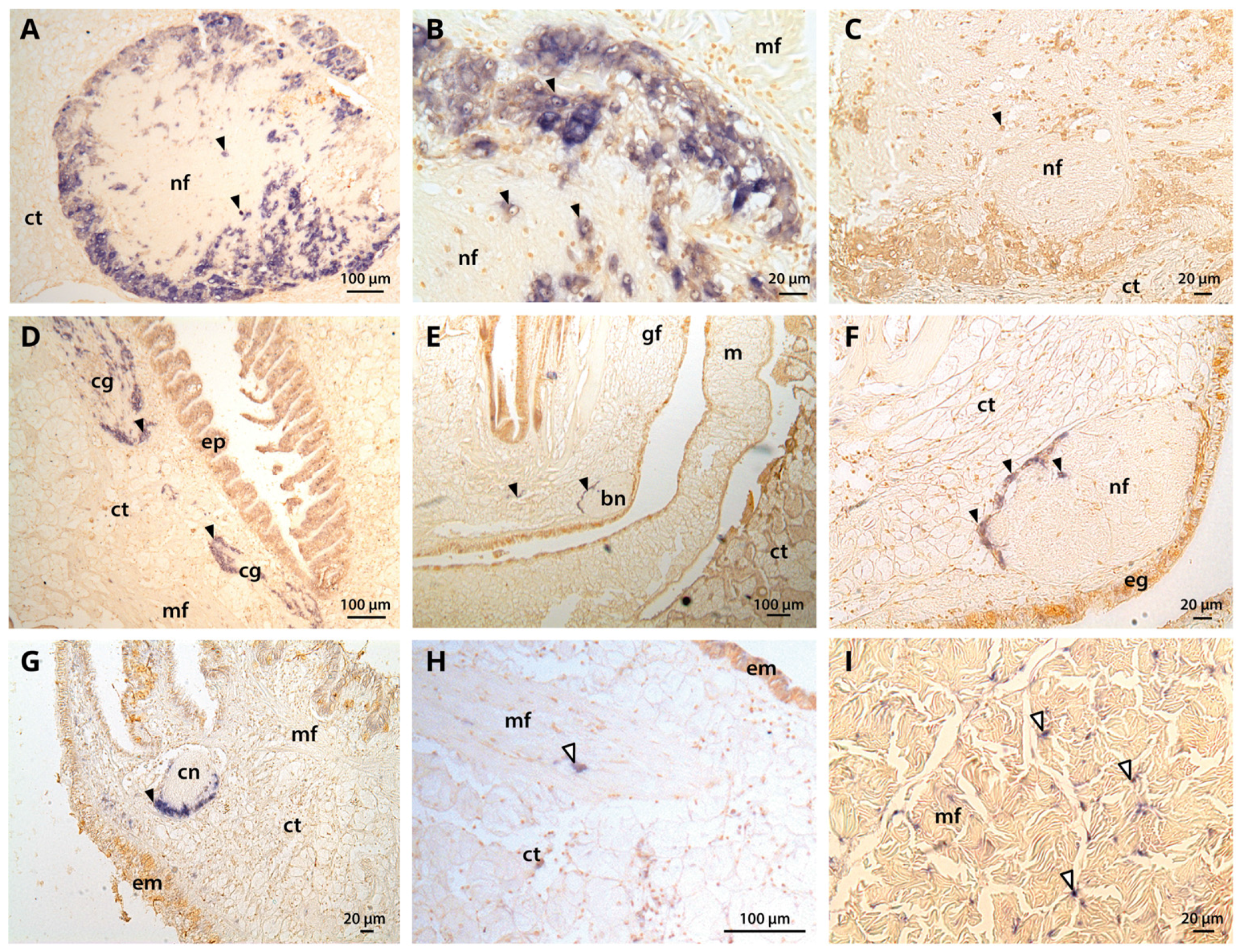
2.5. Tissue-Level CgNav1 α Subunit Expression Patterns
2.6. Expression Patterns of CgNav1 α Subunit Splice Variants
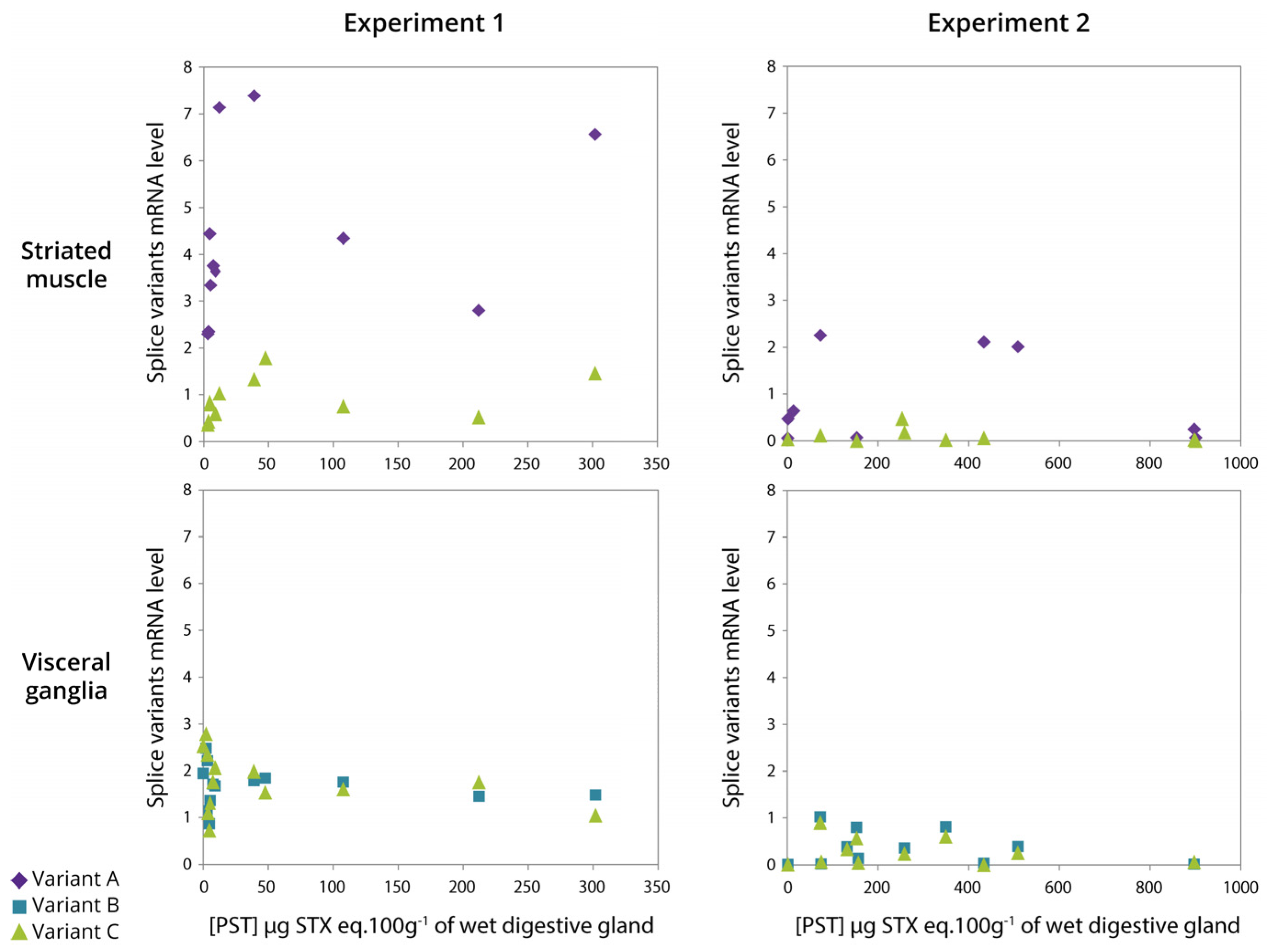
2.7. Relationship between Expression of CgNav1 α Subunit Splice Variants and PST Accumulation
3. Discussion
3.1. Two Genes Encoding Two Types of Nav Channels (Nav1 and Nav2) in C. gigas
3.2. CgNav1 Genotype Could Confer a Certain Resistance of Oysters to PST
3.3. CgNav1 is Spliced in Tissue-Specific Variants
3.4. Potentially Different Pathways of Regulation Exist for CgNav1
3.5. The Level of PST Accumulation Would Be Independent of CgNav1 Transcription Level
4. Materials and Methods
4.1. Phylogenetic Analyses of the Voltage-Gated Sodium Channel α Subunit of Crassostrea gigas
4.2. Biological Material
4.2.1. Crassostrea gigas Oysters
4.2.2. Microalgae Cultures
4.3. Experimental Design for Oyster Exposure to PST
4.3.1. Experiment 1
4.3.2. Experiment 2
4.4. Toxin Quantification by Liquid Chromatography/Fluorescence Detection
4.5. DNA and RNA Extractions and cDNA Synthesis
4.6. Single Nucleotid Polymorphism of C. gigas Nav1 α Subunit Gene
4.7. Amplification and Sequencing of the cDNA of CgNav1 α Subunit
4.8. Localization of CgNav1 α Subunit mRNA Expression by In Situ Hybridization
4.9. Expression of CgNav1 mRNA by Real-Time PCR
4.10. Data Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Van Dolah, F.M. Marine algal toxins: Origins, health effects, and their increased occurrence. Environ. Health Perspect. 2000, 108, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Manfrin, C.; De Moro, G.; Torboli, V.; Venier, P.; Pallavicini, A.; Gerdol, M. Physiological and molecular responses of bivalves to toxic dinoflagellates. Invertebr. Surviv. J. 2012, 9, 184–199. [Google Scholar]
- Sellner, K.G.; Doucette, G.J.; Kirkpatrick, G.J. Harmful algal blooms: Causes, impacts and detection. J. Ind. Microbiol. Biotechnol. 2003, 30, 383–406. [Google Scholar] [CrossRef] [PubMed]
- Bricelj, V.M.; Connell, L.; Konoki, K.; MacQuarrie, S.P.; Scheuer, T.; Catterall, W.A.; Trainer, V.L. Sodium channel mutation leading to saxitoxin resistance in clams increases risk of PSP. Nature 2005, 434, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Catterall, W.A. From ionic currents to molecular mechanisms: The structure and function of voltage-gated sodium channels. Neuron 2000, 26, 13–25. [Google Scholar] [CrossRef]
- Cestèle, S.; Catterall, W.A. Molecular mechanisms of neurotoxin action on voltage-gated sodium channels. Biochimie 2000, 82, 883–892. [Google Scholar] [CrossRef]
- Zakon, H.H. Adaptive evolution of voltage-gated sodium channels: The first 800 million years. Proc. Natl. Acad. Sci. USA 2012, 109, 10619–10625. [Google Scholar] [CrossRef] [PubMed]
- Catterall, W.A. Structure and function of voltage-gated ion channels. Annu. Rev. Biochem. 1995, 64, 493–531. [Google Scholar] [CrossRef] [PubMed]
- Chopra, S.S.; Watanabe, H.; Zhong, T.P.; Roden, D.M. Molecular cloning and analysis of zebrafish voltage-gated sodium channel beta subunit genes: Implications for the evolution of electrical signaling in vertebrates. BMC Evol. Biol. 2007, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Deak, P.; Chopra, M.; Hall, L.M. Cloning and functional analysis of TipE, a novel membrane protein that enhances drosophila para sodium channel function. Cell 1995, 82, 1001–1011. [Google Scholar] [CrossRef]
- Gurnett, C.A.; Campbell, K.P. Transmembrane auxiliary subunits of voltage-dependent ion channels. J. Biol. Chem. 1996, 271, 27975–27978. [Google Scholar] [CrossRef] [PubMed]
- Dong, K. Insect sodium channels and insecticide resistance. Invertebr. Neurosci. 2007, 7, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.H.; Catterall, W.A. Overview of the voltage-gated sodium channel family. Genome Biol. 2003, 4, 1–7. [Google Scholar]
- Anderson, P.A.V.; Roberts-Misterly, J.; Greenberg, R.M. The evolution of voltage-gated sodium channels: Were algal toxins involved? Harmful Algae 2005, 4, 95–107. [Google Scholar] [CrossRef]
- Al-Sabi, A.; McArthur, J.; Ostroumov, V.; French, R.J. Marine toxins that target voltage-gated sodium channels. Mar. Drugs 2006, 4, 157–192. [Google Scholar] [CrossRef]
- Marban, E.; Yamagishi, T.; Tomaselli, G.F. Structure and function of voltage-gated sodium channels. J. Physiol. 1998, 508, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Terlau, H.; Heinemann, S.H.; Stühmer, W.; Pusch, M.; Conti, F.; Imoto, K.; Numa, S. Mapping the site of block by tetrodotoxin and saxitoxin of sodium channel II. FEBS Lett. 1991, 293, 93–96. [Google Scholar] [CrossRef]
- Bricelj, V.M.; MacQuarrie, S.; Twarog, B.M.; Trainer, V.L. Characterization of sensitivity to PSP toxins in North American populations of the softshell clam Mya arenaria. In Harmful Algae 2002; Steidinger, K.A., Landsberg, J.H., Tomas, C.R., Vargo, G.A., Eds.; Florida Institute of Oceanography and Intergovernmental Oceanographic Commission of UNESCO: Paris, France, 2004; pp. 172–174. [Google Scholar]
- Tan, J.; Liu, Z.; Nomura, Y.; Goldin, A.L.; Dong, K. Alternative splicing on an insect sodium channel gene generates pharmacologically distinct sodium channels. J. Neurosci. 2002, 22, 5300–5309. [Google Scholar] [PubMed]
- Haberkorn, H.; Tran, D.; Massabuau, J.C.; Ciret, P.; Savar, V.; Soudant, P. Relationship between valve activity, microalgae concentration in the water and toxin accumulation in the digestive gland of the Pacific oyster Crassostrea gigas exposed to Alexandrium minutum. Mar. Pollut. Bull. 2011, 62, 1191–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Morrison, C.M. The histology and ultrastructure of the adductor muscle of the Eastern oyster Crassostrea virginica. Am. Malacol. Bull. 1993, 10, 25–38. [Google Scholar]
- Sussarellu, R.; Fabioux, C.; Le Moullac, G.; Fleury, E.; Moraga, D. Transcriptomic response of the Pacific oyster Crassostrea gigas to hypoxia. Mar. Genom. 2010, 3, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Riviere, G.; Klopp, C.; Ibouniyamine, N.; Huvet, A.; Boudry, P.; Favrel, P. GigaTON: An extensive publicly searchable database providing a new reference transcriptome in the pacific oyster Crassostrea gigas. BMC Bioinform. 2015, 16, 401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinemann, S.H.; Terlau, H.; Stuhmer, W.; Imoto, K.; Numa, S. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature 1992, 356, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Gur Barzilai, M.; Reitzel, A.M.; Kraus, J.E.M.; Gordon, D.; Technau, U.; Gurevitz, M.; Moran, Y. Convergent Evolution of Sodium Ion Selectivity in Metazoan Neuronal Signaling. Cell Rep. 2012, 2, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Liebeskind, B.J.; Hillis, D.M.; Zakon, H.H. Evolution of sodium channels predates the origin of nervous systems in animals. Proc. Natl. Acad. Sci. USA 2011, 108, 9154–9159. [Google Scholar] [CrossRef] [PubMed]
- Lipkind, G.M.; Fozzard, H.A. Voltage-gated Na+ channel selectivity: The role of the conserved domain III lysine residue. J. Gen. Physiol. 2008, 131, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Schlief, T.; Schönherr, R.; Imoto, K.; Heinemann, H.S. Pore properties of rat brain II sodium channels mutated in the selectivity filter domain. Eur. Biophys. J. 1996, 25, 75–91. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Chung, I.; Liu, Z.; Goldin, A.L.; Dong, K. A voltage-gated calcium-selective channel encoded by a sodium channel-like gene. Neuron 2004, 42, 101–112. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, Z.; Song, W.; Du, Y.; Dong, K. Molecular characterization and functional expression of the DSC1 channel. Insect Biochem. Mol. Biol. 2011, 41, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Du, Y.; Rinkevich, F.; Wang, L.; Xu, P. The Drosophila Sodium Channel 1 (DSC1): The founding member of a new family of voltage-gated cation channels. Pestic. Biochem. Physiol. 2015, 120, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, C.; Bierne, N.; Lapegue, S.; Boudry, P. Single Nucleotide polymorphisms and their relationship to codon usage bias in the Pacific oyster Crassostrea gigas. Gene 2007, 406, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Goldin, A.L. Evolution of voltage-gated Na+ channels. J. Exp. Biol. 2002, 205, 575–584. [Google Scholar] [PubMed]
- Jost, M.C.; Hillis, D.M.; Lu, Y.; Kyle, J.W.; Fozzard, H.A.; Zakon, H.H. Toxin-resistant sodium channels: Parallel adaptive evolution across a complete gene family. Mol. Biol. Evol. 2008, 25, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Kontis, K.J.; Goldin, A.L. Site-directed mutagenesis of the putative pore region of the rat IIA sodium channel. Mol. Pharmacol. 1993, 43, 635–644. [Google Scholar] [PubMed]
- Thackeray, J.R.; Ganetzky, B. Developmentally regulated alternative splicing generates a complex array of Drosophila para sodium channel isoforms. J. Neurosci. 1994, 14, 2569–2578. [Google Scholar] [PubMed]
- Lin, W.H.; Wright, D.E.; Muraro, N.I.; Baines, R.A. Alternative splicing in the voltage-gated sodium channel DmNav regulates activation, inactivation, and persistent current. J. Neurophysiol. 2009, 102, 1994–2006. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Ingles, P.J.; Knipple, D.C.; Soderlund, D.M. Developmental regulation of alternative exon usage in the house fly Vssc1 sodium channel gene. Invertebr. Neurosci. 2002, 4, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.M.; Dong, K.; Tang, Z.H.; Zhang, C.X. Molecular characterization of a sodium channel gene from the Silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2009, 39, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Dai, A.; Temporal, S.; Schulz, D.J. Cell-specific patterns of alternative splicing of voltage-gated ion channels in single identified neurons. Neuroscience 2010, 168, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Schaller, K.L.; Krzemien, D.M.; McKenna, N.M.; Caldwell, J.H. Alternatively spliced sodium channel transcripts in brain and muscle. J. Neurosci. 1992, 12, 1370–1381. [Google Scholar] [PubMed]
- Plummer, N.W.; Galt, J.; Jones, J.M.; Burgess, D.l.; Sprunger, L.K.; Kohrman, D.C.; Meisler, M.H. Exon organization, coding sequence, physical mapping, and polymorphic intragenic markers for the human neuronal sodium channel gene SCN8A. Genomics 1998, 54, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Awad, S.S.; Lightowlers, R.N.; Young, C.; Chrzanowska-Lightowlers, Z.M.; Lømo, T.; Slater, C.R. Sodium channel mRNAs at the neuromuscular junction: Distinct patterns of accumulation and effects of muscle activity. J. Neurosci. 2001, 21, 8456–8463. [Google Scholar] [PubMed]
- Smith, R.D.; Goldin, A.L. Phosphorylation of brain sodium channels in the I-II linker modulates channel function in Xenopus oocytes. J. Neurosci. 1996, 16, 1965–1974. [Google Scholar] [PubMed]
- Lavialle-Defaix, C.; Gautier, H.; Defaix, A.; Lapied, B.; Grolleau, F. Differential regulation of two distinct voltage-dependent sodium currents by group III metabotropic glutamate receptor activation in insect pacemaker neurons. J. Neurophysiol. 2006, 96, 2437–2450. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Ikeda, T.; Salgado, V.L.; Yeh, J.Z.; Narahashi, T. Block of two subtypes of sodium channels in coackroch neurons by indoxacarb insecticides. Neuro Toxicol. 2005, 26, 455–465. [Google Scholar]
- Shiraishi, S.; Yokoo, H.; Yanagita, T.; Kobayashi, H.; Minami, S.-I.; Saitoh, T.; Takasaki, M.; Wada, A. Differential effects of bupivacaine enantiomers, ropivacaine and lidocaine on up-regulation of cell surface voltage-dependent sodium channels in adrenal chromaffin cells. Brain Res. 2003, 966, 175–184. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. Tree Figure Drawing Tool, version 1.4.2; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2007. [Google Scholar]
- Anderson, P.A.; Holman, M.A.; Greenberg, R.M. Deduced amino acid sequence of a putative sodium channel from the scyphozoan jellyfish Cyanea capillata. Proc. Natl. Acad. Sci. USA 1993, 90, 7419–7423. [Google Scholar] [CrossRef] [PubMed]
- Spafford, J.D.; Spencer, A.N.; Gallin, W.J. Genomic organization of a voltage-gated Na+ channel in a hydrozoan jellyfish: Insights into the evolution of voltage-gated Na+ channel genes. Recept. Channels 1999, 6, 493–506. [Google Scholar] [PubMed]
- White, G.B.; Pfahnl, A.; Haddock, S.; Lamers, S.; Greenberg, R.M.; Anderson, P.A. Structure of a putative sodium channel from the sea anemone Aiptasia pallida. Invertebr. Neurosci. 1998, 3, 317–326. [Google Scholar] [CrossRef]
- Liu, Z.; Chung, I.; Dong, K. Alternative splicing of the BSC1 gene generates tissue-specific isoforms in the German cockroach. Insect Biochem. Mol. Biol. 2001, 31, 703–713. [Google Scholar] [CrossRef]
- Zhan, S.; Merlin, C.; Boore, J.L.; Reppert, S.M. The monarch butterfly genome yields insights into long-distance migration. Cell 2011, 147, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Salkoff, L.; Butler, A.; Wei, A.; Scavarda, N.; Giffen, K.; Ifune, C.; Goodman, R.; Mandel, G. Genomic organization and deduced amino acid sequence of a putative sodium channel gene in Drosophila. Science 1987, 237, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Kaneko, Y.; Matsumoto, G.; Hanyu, Y. Cloning and distribution of a putative tetrodotoxin-resistant Na+ channel in newt retina. Zool. Sci. 1999, 16, 587–594. [Google Scholar] [CrossRef]
- Rosenthal, J.J.; Gilly, W.F. Amino acid sequence of a putative sodium channel expressed in the giant axon of the squid Loligo opalescens. Proc. Natl. Acad. Sci. USA 1993, 90, 10026–10030. [Google Scholar] [CrossRef] [PubMed]
- Dyer, J.R.; Johnston, W.L.; Castellucci, V.F.; Dunn, R.J. Cloning and tissue distribution of the Aplysia Na+ channel α-subunit cDNA. DNA Cell Biol. 1997, 16, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Schulz, D.J.; Goaillard, J.-M.; Marder, E.E. Quantitative expression profiling of identified neurons reveals cell-specific constraints on highly variable levels of gene expression. Proc. Natl. Acad. Sci. USA 2007, 104, 13187–13191. [Google Scholar] [CrossRef] [PubMed]
- Dong, K. A single amino acid change in the para sodium channel protein is associated with knockdown-resistance (kdr) to pyrethroid insecticides in German cockroach. Insect Biochem. Mol. Biol. 1997, 27, 93–100. [Google Scholar] [CrossRef]
- Loughney, K.; Kreber, R.; Ganetzky, B. Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell 1989, 58, 1143–1154. [Google Scholar] [CrossRef]
- Chang, C.; Shen, W.K.; Wang, T.T.; Lin, Y.H.; Hsu, E.L.; Dai, S.M. A novel amino acid substitution in a voltage-gated sodium channel is associated with knockdown resistance to permethrin in Aedes aegypti. Insect Biochem. Mol. Biol. 2009, 39, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Kamiyama, T.; Yamauchi, H.; Nagai, S.; Yamaguchi, M. Differences in abundance and distribution of Alexandrium cysts in Sendai Bay, northern Japan, before and after the tsunami caused by the Great East Japan Earthquake. J. Oceanogr. 2014, 70, 185–195. [Google Scholar] [CrossRef]
- Guillard, R.R.L.; Hargraves, P.E. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia 1993, 32, 234–236. [Google Scholar] [CrossRef]
- Van de Riet, J.; Gibbs, R.S.; Muggah, P.M.; Rourke, W.A.; MacNeil, J.D.; Quilliam, M.A. Liquid Chromatography Post-Column Oxidation (PCOX) Method for the Determination of Paralytic Shellfish Toxins in Mussels, Clams, Oysters, and Scallops: Collaborative Study. J. AOAC Int. 2011, 94, 1154–1176. [Google Scholar] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Howard, D.W.; Lewis, E.J.; Keller, B.J.; Smith, C.S. Histological Techniques for Marine Bivalve Mollusks and Crustaceans, 2nd ed.NOAA: Oxford, MD, 2004; p. 218.
- Santerre, C.; Sourdaine, P.; Martinez, A.S. Expression of a natural antisense transcript of Cg-Foxl2 during the gonadic differentiation of the oyster Crassostrea gigas: First demonstration in the gonads of a lophotrochozoa species. Sex. Dev. Genet. Mol. Biol. Evol. Endocrinol. Embryol. Pathol. Sex Determ. Differ. 2012, 6, 210–221. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.T.K.; Tan, T.W.; Ranganathan, S. MGAlignIt: A web service for the alignment of mRNA/EST and genomic sequences. Nucleic Acids Res. 2003, 31, 3533–3536. [Google Scholar] [CrossRef] [PubMed]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Callebaut, I.; Labesse, G.; Durand, P.; Poupon, A.; Canard, L.; Chomilier, J.; Henrissat, B.; Mornon, P.J. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): Current status and perspectives. Cell. Mol. Life Sci. CMLS 1997, 53, 621–645. [Google Scholar] [CrossRef] [PubMed]
- Blom, N.; Sicheritz-Pontén, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.-H.; Lee, T.-Y.; Liang, H.-K.; Huang, C.-M.; Wang, T.-Y.; Yang, Y.-H.; Chu, C.-H.; Huang, H.-D.; Ko, M.-T.; Hwang, J.-K. KinasePhos 2.0: A web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res. 2007, 35, W588–W594. [Google Scholar] [CrossRef] [PubMed]
- Neuberger, G.; Schneider, G.; Eisenhaber, F. pkaPS: Prediction of protein kinase A phosphorylation sites with the simplified kinase-substrate binding model. Biol. Direct 2007, 2, 1–23. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]



| Domains | Populations | N | Pe | Pi | Le | Li | ∏e | ∏i | ∏t |
|---|---|---|---|---|---|---|---|---|---|
| DI | LOG | 15 | 1 | 3 | 117 | 311 | 0.009 | 0.010 | 0.009 |
| LB | 12 | 1 | 2 | 117 | 311 | 0.009 | 0.060 | 0.007 | |
| RE | 20 | 1 | 3 | 117 | 311 | 0.009 | 0.010 | 0.009 | |
| JAP | 13 | 1 | 3 | 117 | 311 | 0.009 | 0.010 | 0.009 | |
| DII | LOG | 48 | 5 | - | 136 | - | 0.037 | - | 0.037 |
| LB | 46 | 5 | - | 136 | - | 0.037 | - | 0.037 | |
| RE | 42 | 5 | - | 136 | - | 0.037 | - | 0.037 | |
| JAP | 19 | 6 | - | 136 | - | 0.044 | - | 0.044 | |
| DIII | LOG | 14 | 0 | 7 | 140 | 188 | 0 | 0.037 | 0.021 |
| LB | 8 | 0 | 6 | 140 | 188 | 0 | 0.032 | 0.018 | |
| RE | 11 | 0 | 5 | 140 | 188 | 0 | 0.027 | 0.015 | |
| JAP | 7 | 0 | 5 | 140 | 188 | 0 | 0.027 | 0.015 | |
| DIV | LOG | 26 | 4 | - | 301 | - | 0.013 | - | 0.013 |
| LB | 25 | 5 | - | 301 | - | 0.017 | - | 0.017 | |
| RE | 25 | 5 | - | 301 | - | 0.017 | - | 0.017 | |
| JAP | 20 | 4 | - | 301 | - | 0.013 | - | 0.013 |
| Name of Sequences | Total cDNA Size (bp) | Total Predicted Protein Size (aa) | Alternatively Spliced Fragments | Spliced cDNA Fragments Size (bp) | Spliced Protein Fragments Size (aa) |
|---|---|---|---|---|---|
| CGI_10001852 | 5205 | 1734 | / | / | / |
| Variant A | 5532 | 1844 | −exon 7 | −405 | −135 |
| +exon 15 | +33 | +11 | |||
| Variant B | 5877 | 1959 | −exon 5 | −27 | −9 |
| Variant C | 5505 | 1835 | −exons 5 and 7 | −432 | −144 |
| +exon 15 | +33 | +11 |
| Amplicon Names | Primer Names | Primer Sequences (5′–3′) | Length (bp) |
|---|---|---|---|
| Primers used for the in situ hybridization | |||
| Exon 24 | CgNav9_e25F | AGGCGGGTGTTATGTTCTTG | 20 |
| CgNav9_e25R | GCGGTATCTTCGTGAATGGT | 20 | |
| Primers used in splice variants real-time PCR | |||
| Variant A | CgNav9_a5F | CTCTTGTGCTCATTCCAGCA | 20 |
| CgNav9_s7R | GACCCATTTATTGACCCCTTCT | 22 | |
| Variant B | CgNav9_s5F | CGAAAGATTCAACAAACAATGCATG | 25 |
| CgNav9_a7R1 | TTAAAGGTTGATGGTCAGCGTGATT | 25 | |
| Variant C | CgNav9_s5F | CGAAAGATTCAACAAACAATGCATG | 25 |
| CgNav9_s7R | GACCCATTTATTGACCCCTTCT | 22 | |
| Variant D | CgNav9_a5F | CTCTTGTGCTCATTCCAGCA | 20 |
| CgNav9_a7R1 | TTAAAGGTTGATGGTCAGCGTGATT | 25 | |
| GAPDH | qFw_GAPDH | GGAGACAAGCGAAGCAGCAT | 20 |
| qRev_GAPDH | CACAAAATTGTCATTCAAGGCAAT | 24 | |
| EF1α | qfElongN | GATTGCCACACTGCTCACAT | 20 |
| qrElongN | AGCATCTCCGTTCTTGATGC | 20 | |
| Species Name | Nav Name | Common Name | GenBank Accession Number | Size (aa) | Reference |
|---|---|---|---|---|---|
| Cyanea capillata | CcNav | Lion’s mane jellyfish | AAA75572 | 1740 | [54] |
| Polyorchis penicillatus | PpNav | Hydrozoan jellyfish | AAC09306 | 1695 | [55] |
| Aiptasia pallida | ApNav | Pale anemone | AAB96953 | 1810 | [56] |
| Capitella teleta | CtNav2 | Polychaete worm | JGI 134859 * | 1694 | |
| Crassostrea gigas | CgNav2 (Nav5) | Pacific oyster | EKC21550 | 1986 | [21] |
| Blattella germanica | BgNav2 (BSC1) | German cockroach | AAK01090 | 2304 | [57] |
| Danaus plexippus | DpNav2 | Monarch butterfly | EHJ64356 | 1991 | [58] |
| Drosophila melanogaster | DmNav2 (DSC1) | Fruit fly | ABF70206 | 2409 | [59] |
| Cynops pyrrhogaster | CpNav | Japanese common newt | AAD17315 | 2007 | [60] |
| Capitella teleta | CtNav1 | Polychaete worm | JGI 210954 * | 1690 | |
| Doryteuthis opalescens | DoNav | Opalescent inshore squid | AAA16202 | 1784 | [61] |
| Aplysia californica | AcNav1 | California sea hare | NP_001191637 | 1993 | [62] |
| Mya arenaria | MaNav | Softshell clam | AAX14719 | 1435 | [4] |
| Crassostrea gigas | CgNav1 (Nav9) | Pacific oyster | EKC22630 | 1734 | [21] |
| Cancer borealis | CbNav | Jonah crab | ABL10360 | 1989 | [63] |
| Blattella germanica | BgNav1 (para) | German cockroach | AAC47483 | 2031 | [64] |
| Danaus plexippus | DpNav1 (para) | Monarch butterfly | EHJ74501 | 2112 | [58] |
| Drosophila melanogaster | DmNav1 (para) | Fruit fly | AAB59195 | 2131 | [65] |
| Aedes aegypti | AaNav1 (para) | Yellow fever mosquito | ACB37023 | 2140 | [66] |
| Culex pipiens pallens | CpNav1 (para) | Northern house mosquito | AGO33659 | 2043 | |
| Culex quinquefasciatus | CqNav1 (para) | Southern house mosquito | AGO33660 | 2052 |
| Amplicon Names | Primer Names | Primer Sequences (5′–3′) | Length (bp) | Amplicon Size (bp) |
|---|---|---|---|---|
| Primers used for segment P region amplification | ||||
| DI | CgNav9_1f | TGACACTCACACAAACCCAGA | 21 | 491 |
| CgNav9_1r | AACGAGCCCAGCAGTATCAC | 20 | ||
| DII | CgNav9_2f | TGTTCTTGCCATTGTGGTGT | 20 | 214 |
| CgNav9_2r | AAAGAACGGGACACAGATCG | 20 | ||
| DIII | CgNav9_3f’2 | GGTGTGCCTCATTTTCTGGT | 20 | 385 |
| CgNav9_3r’2 | CTGCACCGATATTCTCAGCA | 20 | ||
| DIV | CgNav9_4f | GACGTCATGGACCAATTCCT | 20 | 353 |
| CgNav9_4r | TTACAACCCTCCTCGTTCGT | 20 | ||
| Primers used for the amplification of full length cDNA | ||||
| a | CgNav9_TF2 | GCTGTGTACGACTAAAATGGAG | 22 | 425 |
| CgNav9_e1R | ACGCGCTGAATAATGGATG | 19 | ||
| b | CgNav9_Ch2F | AGCCCCTTTAACCCACTCAG | 20 | 863 |
| CgNav9_1R | AACGAGCCCAGCAGTATCAC | 20 | ||
| c | CgNav9_Ch2F | AGCCCCTTTAACCCACTCAG | 20 | 1873 |
| CgNav9_Ch4R | CAAAAGCATCCAACACGATG | 20 | ||
| d | CgNav9_Ch5F | AGCGACTACCTTCCTTTCGAG | 21 | 984 |
| CgNav9_2Rs | GCTTGGTTCTCTCTCGTTCG | 20 | ||
| e | CgNav9_Ch6F | GGAAGATGGGTCAAAGTCAAAG | 22 | 1172 |
| CgNav9_3r’2 | GCGTCATTCATTACTTCGATCC | 22 | ||
| f | CgNav9_Ch8F | CCTGAATCTGTTCATCGGTGT | 21 | 801 |
| CgNav9_4R | TTACAACCCTCCTCGTTCGT | 20 | ||
| g | CgNav9_Ch9F | CACGTTCGGGATGAGTTTCT | 20 | 867 |
| CgNav9_e25R | GCGGTATCTTCGTGAATGGT | 20 | ||
| h | CgNav9_Ch10F | ACTACGCCGCAAGGGTTAT | 19 | 590 |
| CgNav9_TR | GGGTTGATAACAGTGGGTGAA | 21 | ||
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boullot, F.; Castrec, J.; Bidault, A.; Dantas, N.; Payton, L.; Perrigault, M.; Tran, D.; Amzil, Z.; Boudry, P.; Soudant, P.; et al. Molecular Characterization of Voltage-Gated Sodium Channels and Their Relations with Paralytic Shellfish Toxin Bioaccumulation in the Pacific Oyster Crassostrea gigas. Mar. Drugs 2017, 15, 21. https://doi.org/10.3390/md15010021
Boullot F, Castrec J, Bidault A, Dantas N, Payton L, Perrigault M, Tran D, Amzil Z, Boudry P, Soudant P, et al. Molecular Characterization of Voltage-Gated Sodium Channels and Their Relations with Paralytic Shellfish Toxin Bioaccumulation in the Pacific Oyster Crassostrea gigas. Marine Drugs. 2017; 15(1):21. https://doi.org/10.3390/md15010021
Chicago/Turabian StyleBoullot, Floriane, Justine Castrec, Adeline Bidault, Natanael Dantas, Laura Payton, Mickael Perrigault, Damien Tran, Zouher Amzil, Pierre Boudry, Philippe Soudant, and et al. 2017. "Molecular Characterization of Voltage-Gated Sodium Channels and Their Relations with Paralytic Shellfish Toxin Bioaccumulation in the Pacific Oyster Crassostrea gigas" Marine Drugs 15, no. 1: 21. https://doi.org/10.3390/md15010021






