Merosesquiterpene Congeners from the Australian Sponge Hyrtios digitatus as Potential Drug Leads for Atherosclerosis Disease
Abstract
:1. Introduction
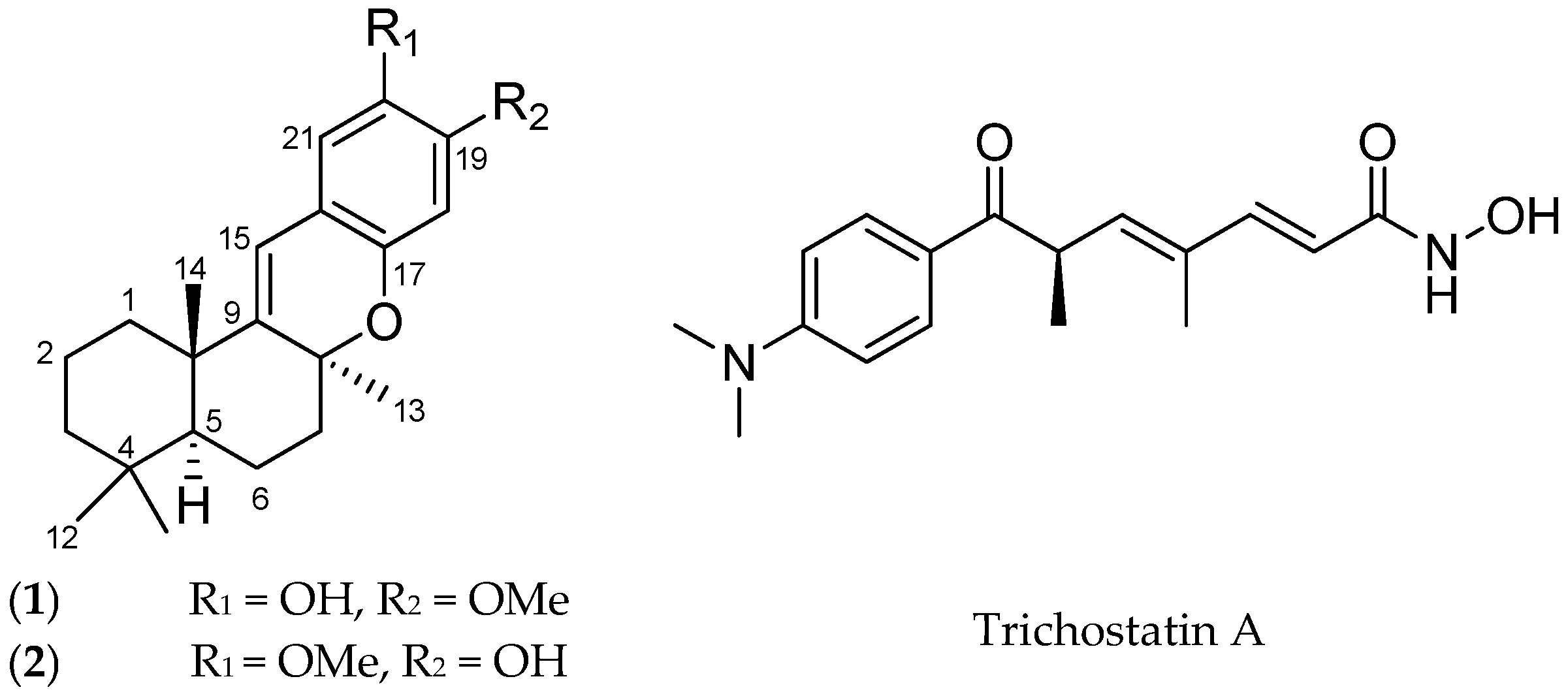
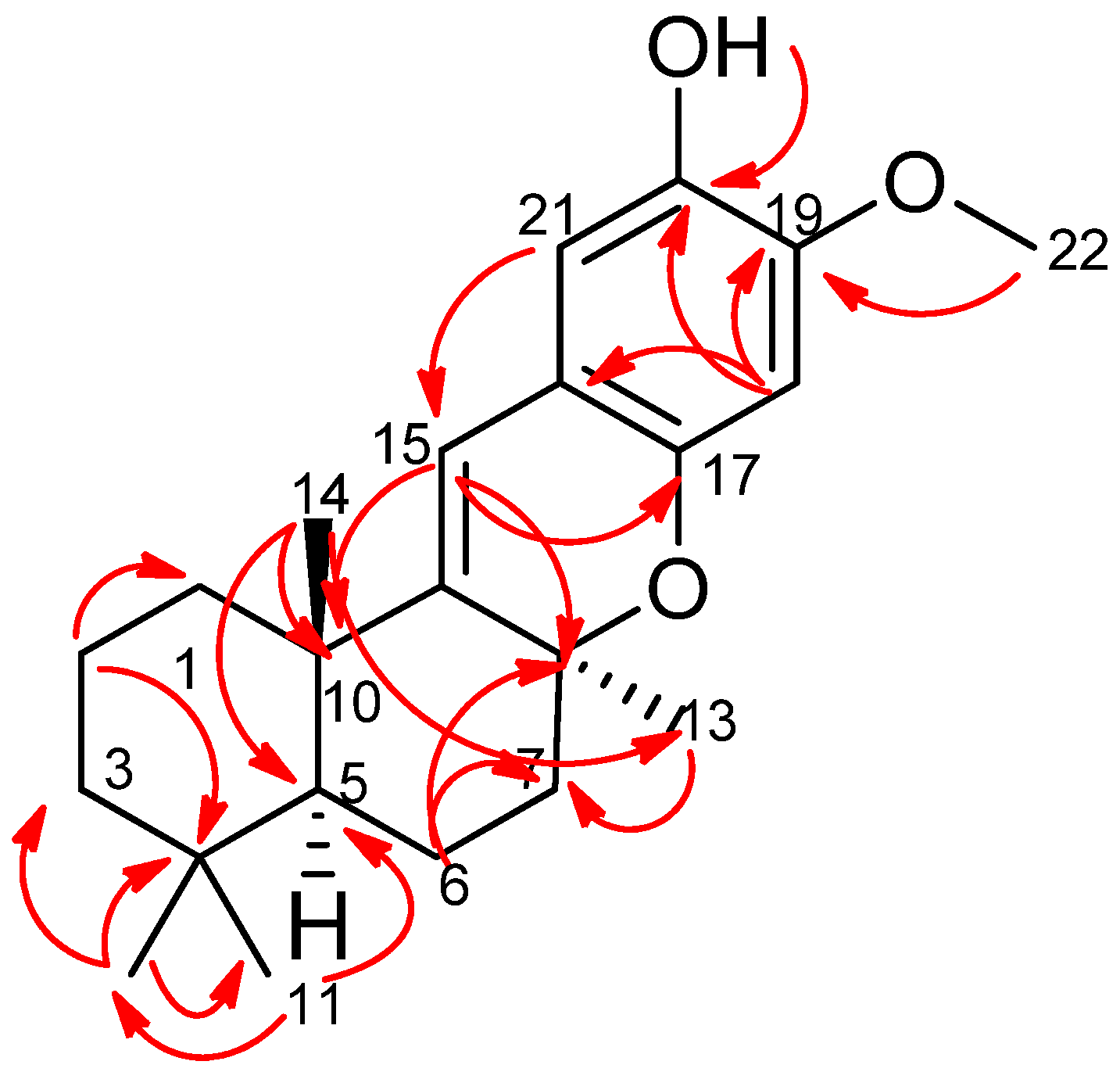
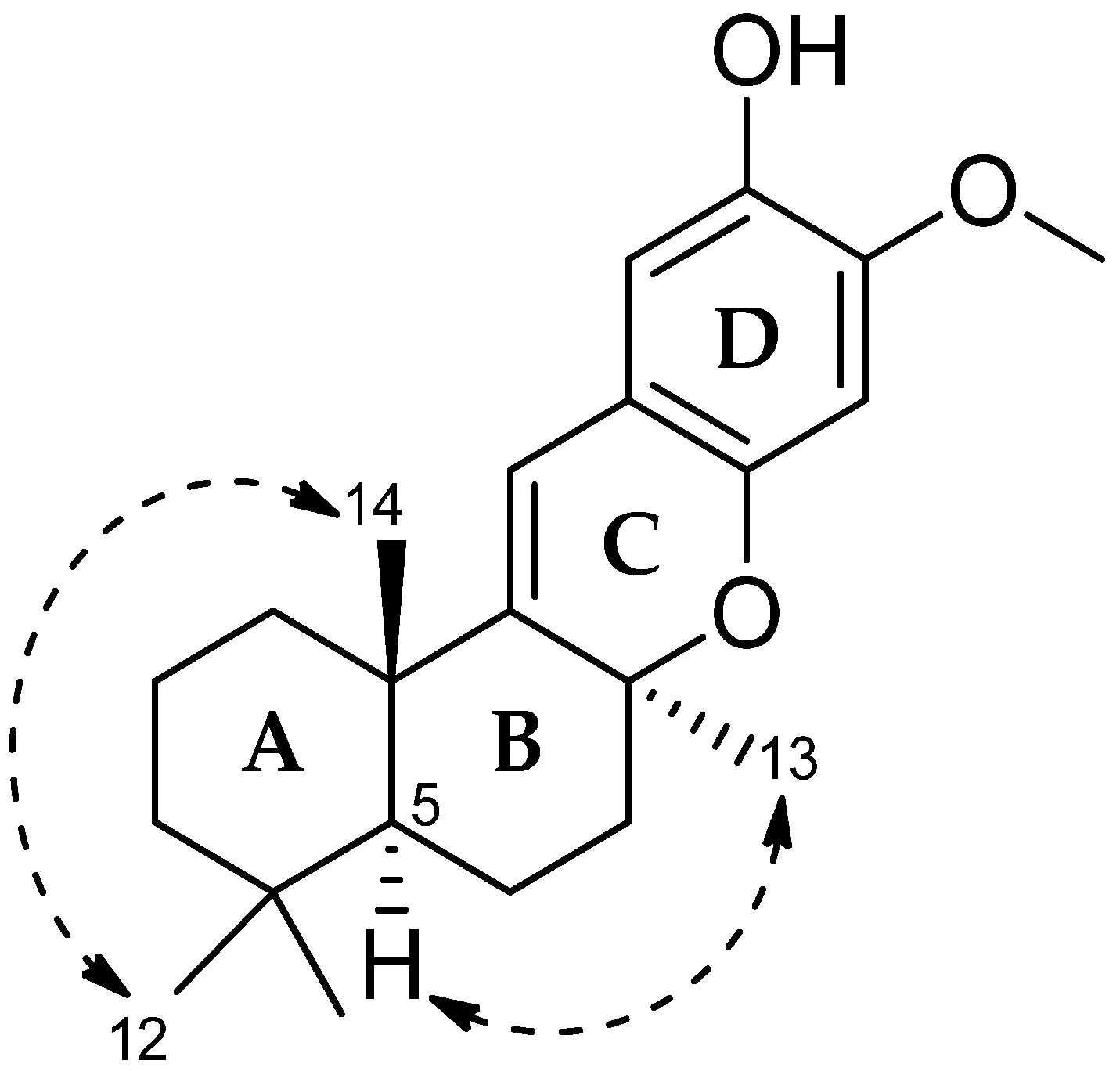
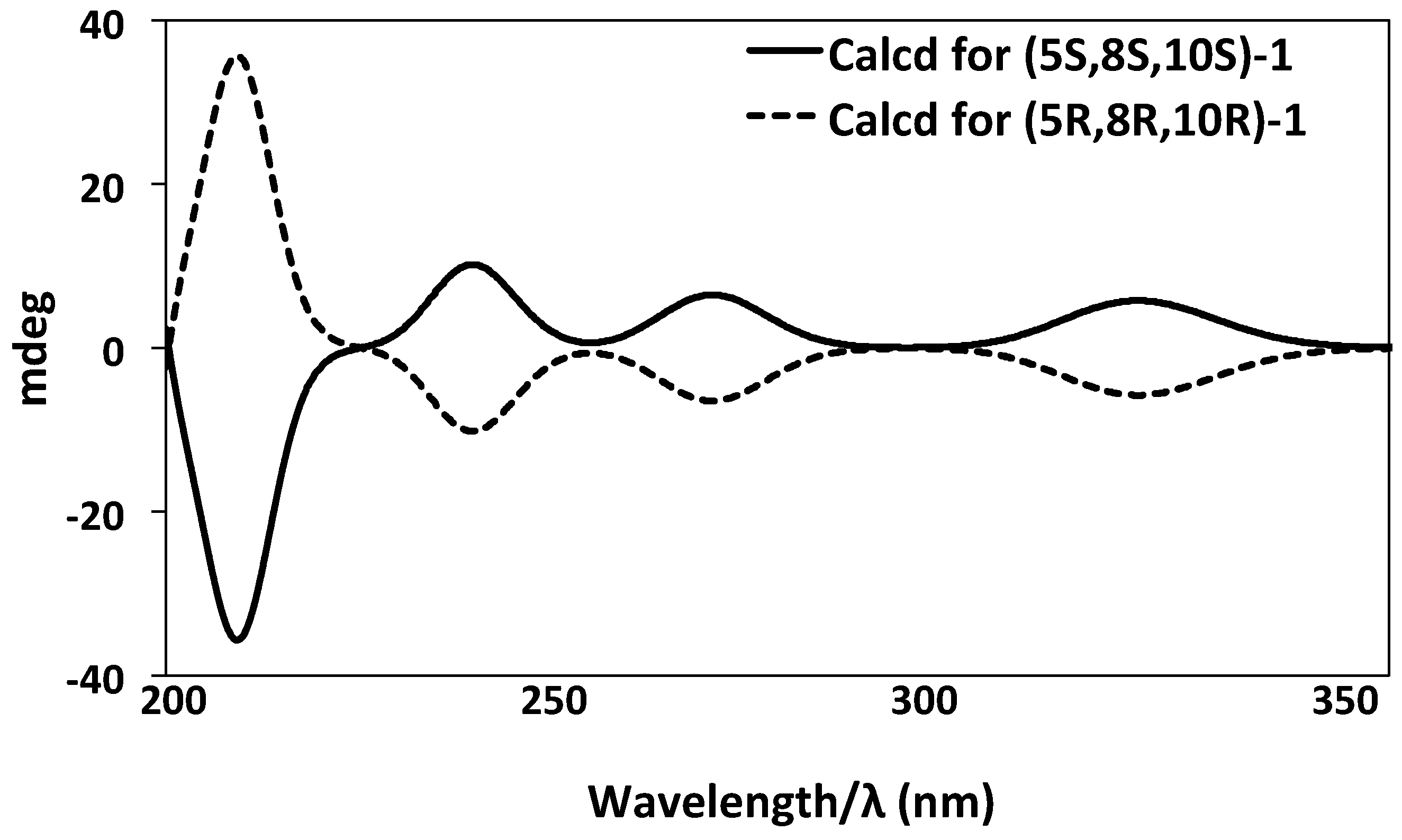
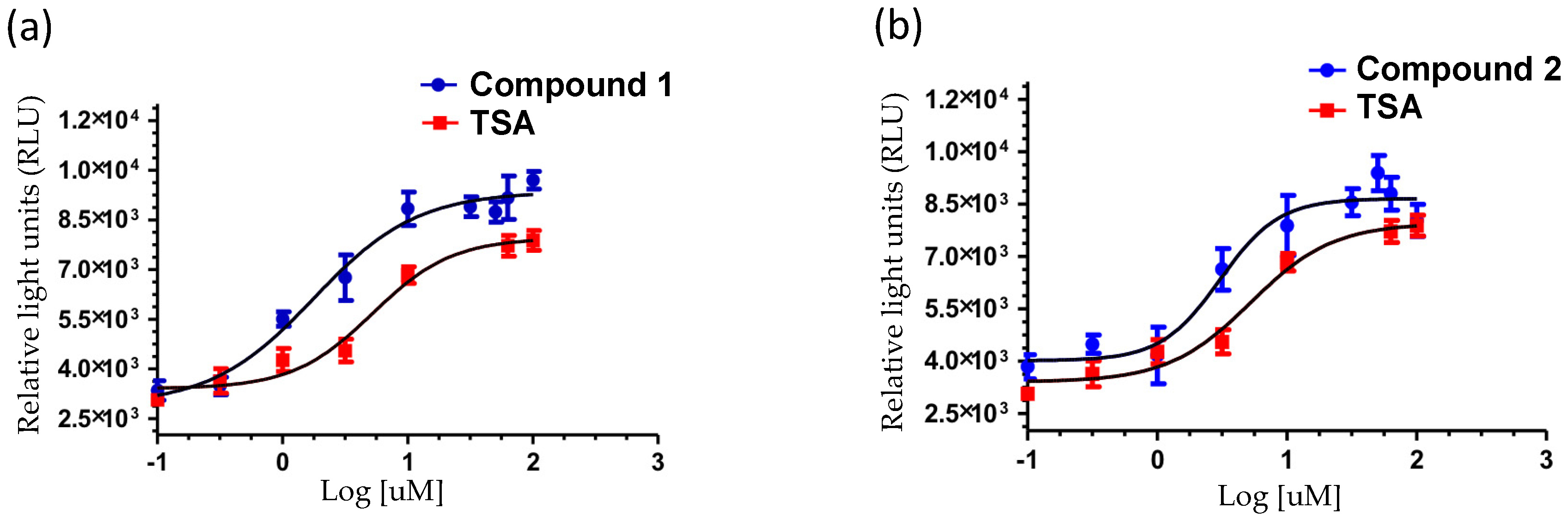
2. Results and Discussion
3. Materials and Methods
3.1. General
3.2. Animal Material
3.3. Extractions and Isolation
3.4. ECD Calculation
3.5. Atherosclerosis Assays
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009, 8, 69–85. [Google Scholar] [CrossRef] [PubMed]
- MarinLit. A Database Marine Natural Products Literature. Available online: http://pubs.rsc.org/marinlit/ (accessed on 30 November 2015).
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2014, 33, 382–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ana, M.; Helena, V.; Helena, G.; Susana, S. Marketed Marine Natural Products in the Pharmaceutical and Cosmeceutical Industries: Tips for Success. Mar Drugs 2014, 12, 1066–1101. [Google Scholar]
- Meyer, C.A. The Global Marine Pharmaceuticals Pipeline. Available online: http://marinepharmacology.midwestern.edu/ (accessed on 22 January 2016).
- De Rosa, S.; de Giulio, A.; Iodice, C. Biological effects of prenylated hydroquinones: Structure–activity relationship studies in antimicrobial, brine shrimp, and fish lethality assays. J. Nat. Prod. 1994, 57, 1711–1716. [Google Scholar] [CrossRef] [PubMed]
- Hamann, M.T.; Scheuer, P.J.; Kelly-Borges, M. Biogenetically diverse, bioactive constituents of a sponge, order Verongida: Bromotyramines and sesquiterpene-shikimate derived metabolites. J. Org. Chem. 1993, 58, 6565–6569. [Google Scholar] [CrossRef]
- Alvi, K.A.; Diaz, M.C.; Crews, P.; Slate, D.L.; Lee, R.H.; Moretti, R. Evaluation of new sesquiterpene quinones from two Dysidea sponge species as inhibitors of protein tyrosine kinase. J. Org. Chem. 1992, 57, 6604–6607. [Google Scholar] [CrossRef]
- Muller, W.E.G.; Maidhof, A.; Zahn, R.K.; Schroder, H.C.; Gasic, M.J.; Heidemann, D.; Bernd, A.; Kurelec, B.; Eich, E.; Seibert, G. Potent antileukemic activity of the novel cytostatic agent avarone and its analogs in vitro and in vivo. Cancer Res. 1985, 45, 4822–4826. [Google Scholar] [PubMed]
- Kondracki, M.L.; Guyot, M. Biologically active quinone and hydroquinone sesquiterpenoids from the sponge Smenospongia sp. Tetrahedron 1989, 45, 1995–2004. [Google Scholar] [CrossRef]
- Hooper, J.N.A.; van Soest, R.W.M. Systema Porifera a Guide to the Classification of Sponges; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2002; Volume 1, pp. 1028–1050. [Google Scholar]
- Youssef, D.T.A.; Shaala, L.A.; Emara, S. Antimycobacterial scalarane-based sesterterpenes from the red sea sponge Hyrtios erecta. J. Nat. Prod. 2005, 68, 1782–1784. [Google Scholar] [CrossRef] [PubMed]
- Youssef, D.T.A.; Singab, A.B.; van Soest, R.W.M.; Fusetani, N. Hyrtiosenolides A and B, two new sesquiterpene γ-methoxybutenolides and a new sterol from a red sea sponge Hyrtios species. J. Nat. Prod. 2004, 67, 1736–1739. [Google Scholar] [CrossRef] [PubMed]
- Salmoun, M.; Devijver, C.; Daloze, D.; Braekman, J.-C.; Gomez, R.; de Kluijver, M.; van Soest, R.W.M. New sesquiterpenes/quinones from two sponges of the genus Hyrtios. J. Nat. Prod. 2000, 63, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Slevin, M.; Matou, S.; Ahmed, N.; Choudhary, M.I.; Ranjit, R.; West, D.; Gaffney, J. Anti-angiogenic activity of sesterterpenes; natural product inhibitors of FGF-2-induced angiogenesis. Angiogenesis 2008, 11, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Aoki, S.; Sakai, H.; Kawazoe, K.; Kihara, N.; Sasaki, T.; Kitagawa, I. Altohyrtins B and C and 5-desacetylalthyrtin A, potent cytotoxic macrolide congeners of altohyrtin A, from the Okinawan marine sponge Hyrtios altum. Chem. Pharm. Bull. 1993, 41, 989–991. [Google Scholar] [CrossRef] [PubMed]
- Youssef, D.T.A. Hyrtioerectines A–C, cytotoxic alkaloids from the red sea sponge Hyrtios erectus. J. Nat. Prod. 2005, 68, 1416–1419. [Google Scholar] [CrossRef] [PubMed]
- Salmoun, M.; Devijver, C.; Daloze, D.; Braekman, J.-C.; van Soest, R.W.M. 5-Hydroxytryptamine-derived alkaloids from two marine sponges of the genus Hyrtios. J. Nat. Prod. 2002, 65, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Aoki, S.; Ye, Y.; Higuchi, K.; Takashima, A.; Tanaka, Y.; Kitagawa, I.; Kobayashi, M. Novel neuronal nitric oxide synthase (nNOS) selective inhibitor, aplysinopsin-type indole alkaloid, from marine sponge Hyrtios erecta. Chem. Pharm. Bull. 2001, 49, 1372–1374. [Google Scholar] [CrossRef] [PubMed]
- Cimino, G.; De Stefano, S.; Minale, L.; Trivellone, E. 12-epi-Scalarin and 12-epi-deoxoscalarin, sesterterpenes from the sponge Spongia nitens. J. Chem. Soc. Perkin Trans. 1977, 13, 1587–1593. [Google Scholar] [CrossRef]
- Utkina, N.K.; Denisenko, V.A.; Krasokhin, V.B. Sesquiterpenoid aminoquinones from the marine sponge Dysidea sp. J. Nat. Prod. 2010, 73, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.E.; Mauricio, G.I.; Alejandro, F.B.; Nelida, S.T.; Ramon, M.C.; Miguel, A.M.; Ana, R.Q. Study of puupehenone and related compounds as inhibitors of angiogenesis. Int. J. Cancer 2004, 110, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Ivette, C.P.; Miranda, L.S.; Philip, C. Puupehenone congeners from an Indo-Pacific Hyrtios sponge. J. Nat. Prod. 2003, 66, 2–6. [Google Scholar]
- Using the Schrödinger API. Available online: https://www.schrodinger.com/docs/suite2015-3/python_api/api/schrodinger.application.macromodel.utils.ComUtilAppSci–class.html (accessed on 15 January 2016).
- Arai, T.; Wang, N.; Bezouevski, M.; Welch, C.; Tall, A.R. Decreased atherosclerosis in heterozygous low density lipoprotein receptor-deficient mice expressing the scavenger receptor B1 transgene. J. Biol. Chem. 1999, 274, 2366–2371. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; van Eck, M.; Van Craeyveld, E.; Jacobs, F.; Carlier, V.; Van Linthout, S.; Erdel, M.; Tjwa, M.; De Geest, B. Critical role of scavenger receptor-BI-expressing bone marrow-derived endothelial progenitor cells in the attenuation of allograft vasculopathy after human Apo A-I transfer. Blood 2009, 113, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Covey, S.D.; Krieger, M.; Wang, W.; Penman, M.; Trigatti, B.L. Scavenger receptor class B type 1-mediated protection against atherosclerosis in LDL receptor-negative mice involves its expression in bone marrow-derived cells. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 1589–1594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Patricia, G.Y.; Yan, R.S.; Vladimir, R.B.; Yuomin, Z.; Sergio, F.; MacRae, F.L. Inactivation of macrophage scavenger receptor class B type 1 promotes atherosclerotic lesion development in apolipoprotein e-deficient mice. Circulation 2003, 108, 2258–2263. [Google Scholar] [CrossRef] [PubMed]
- Drug Likeness Tools (Drulito). Available online: http://www.niper.ac.in/pi_dev_tools/DruLiToWeb/DruLiTo_index.html (accessed on 28 January 2016).
- Ingo, M. Selection Criteria for Drug–Like Compounds. Med. Res. Rev. 2003, 23, 302–321. [Google Scholar]
- Daniel, F.V.; Stephen, R.J.; Hung, Y.C.; Brian, R.S.; Keith, W.W.; Kenneth, D.K. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar]
- Maestro, version 9.9; Schrödinger, LLC.: New York, NY, USA, 2012.
- Maestro, version 9.3.5; Schrödinger, LLC.: New York, NY, USA, 2012.
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision E.01; Gaussian, Inc.: Wallingford, CT, USA, 2009.


| Position | 1 | 2 | ||||
|---|---|---|---|---|---|---|
| δC | δH | gHMBC b | δC | δH | gHMBC b | |
| 1 | 38.7 | 1.22, m | C-2, 14 | 38.6 | 1.21, m | C-14 |
| 2 | 18.9 | 1.49, m 1.60, m | C-4, 10, 3 C-1, 3 | 18.6 | 1.49, m 1.60, m | C-4 |
| 3 | 41.7 | 1.12, m 1.39, m | C-2, 12, 11 C-2, 5 | 41.6 | 1.11, m 1.40, m | C-12, 5 |
| 4 | 32.6 | 33.4 | ||||
| 5 | 43.4 | 1.37, m | C-4, 6, 12, 14, 9, 10 | 43.5 | 1.38, m | C-6, 12, 14, 4, 10, 9 |
| 6 | 16.7 | 1.64, m 1.76, m | C-7, 4, 5, 8 C-7, 10, 5 | 16.8 | 1.65, m 1.76, m | C-4, 5 C-7, 10 |
| 7 | 30.6 | 1.88, m 2.04, m | C-6, 14, C-6, 14, | 30.7 | 1.88, m 2.05, m | C-6, 13, 8, 5, 9 C-6, 13, 3, 8 |
| 8 | 75.9 | 75.9 | ||||
| 9 | 148.7 | 148.5 | ||||
| 10 | 38.1 | 38.1 | ||||
| 11 | 33.4 | 0.85, s | C-12, 3, 5 | 32.5 | 0.85, s | C-4, 12, 3 |
| 12 | 21.6 | 0.92, s | C-4, 3, 5, 11 | 21.1 | 0.92, s | C-4 |
| 13 | 24.9 | 1.25, s | C-7, 8, 9 | 24.9 | 1.25, s | C-7, 8, 9 |
| 14 | 25.2 | 1.14, m | C-1, 5, 8, 9, 10 | 25.4 | 1.14, m | C-10, 5, 9 |
| 15 | 114.1 | 6.11, s | C-10, 8, 21, 16, 17 | 114.0 | 6.11, s | C-14, 10, 8, 21, 16, 17 |
| 16 | 115.0 | 114.8 | ||||
| 17 | 145.9 | 145.5 | ||||
| 18 | 103.9 | 6.22, s | C-16, 20, 17, 19 | 103.9 | 6.22, s | C-16, 17, 20 |
| 19 | 141.9 | 146.6 | ||||
| 19-OH | 8.95, s | C-18, 19, 20 | ||||
| 20 | 146.8 | 141.9 | ||||
| 20-OH | 8.98, s | C-18, 20, 19 | ||||
| 21 | 110.9 | 6.67, s | C-15, 20, 17, 19 | 111.0 | 6.67, s | C-15, 18, 19, 20 |
| 22 | 56.7 | 3.68, s | C-19 | 56.3 | 3.68, s | C-20 |
| Compound | EC50 (μM) b | % Efficacy (E) |
|---|---|---|
| (5S,8S,10S)-19-methoxy-9,15-ene-puupehenol (1) | 1.78 | 130 |
| (5S,8S,10S)-20-methoxy-9,15-ene-puupehenol (2) | 3.05 | 121 |
| Trichostatin A a | 5.25 | 100 |
| Compound | MW | c logP | HBA | HBD | PSA | NROT | Predicted Bioavailability |
|---|---|---|---|---|---|---|---|
| 1 | 342.34 | 5.05 | 3 | 1 | 38.69 | 1 | √ |
| 2 | 342.45 | 5.05 | 3 | 1 | 38.69 | 1 | √ |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wahab, H.A.; Pham, N.B.; Muhammad, T.S.T.; Hooper, J.N.A.; Quinn, R.J. Merosesquiterpene Congeners from the Australian Sponge Hyrtios digitatus as Potential Drug Leads for Atherosclerosis Disease. Mar. Drugs 2017, 15, 6. https://doi.org/10.3390/md15010006
Wahab HA, Pham NB, Muhammad TST, Hooper JNA, Quinn RJ. Merosesquiterpene Congeners from the Australian Sponge Hyrtios digitatus as Potential Drug Leads for Atherosclerosis Disease. Marine Drugs. 2017; 15(1):6. https://doi.org/10.3390/md15010006
Chicago/Turabian StyleWahab, Huda A., Ngoc B. Pham, Tengku S. Tengku Muhammad, John N. A. Hooper, and Ronald J. Quinn. 2017. "Merosesquiterpene Congeners from the Australian Sponge Hyrtios digitatus as Potential Drug Leads for Atherosclerosis Disease" Marine Drugs 15, no. 1: 6. https://doi.org/10.3390/md15010006





