High-Throughput Identification of Antimicrobial Peptides from Amphibious Mudskippers
Abstract
:1. Introduction
2. Results
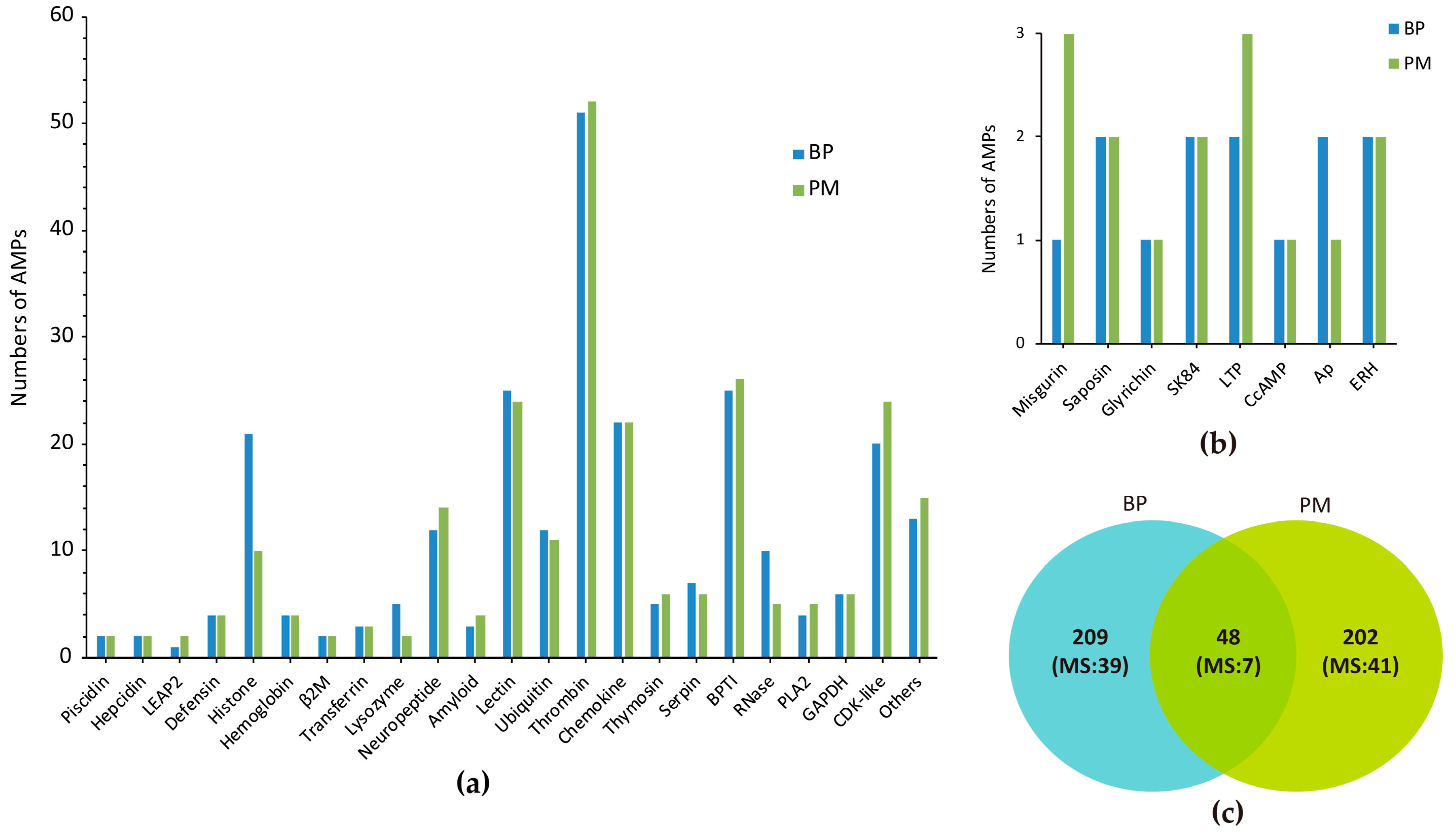
2.1. Summary of Achieved Data
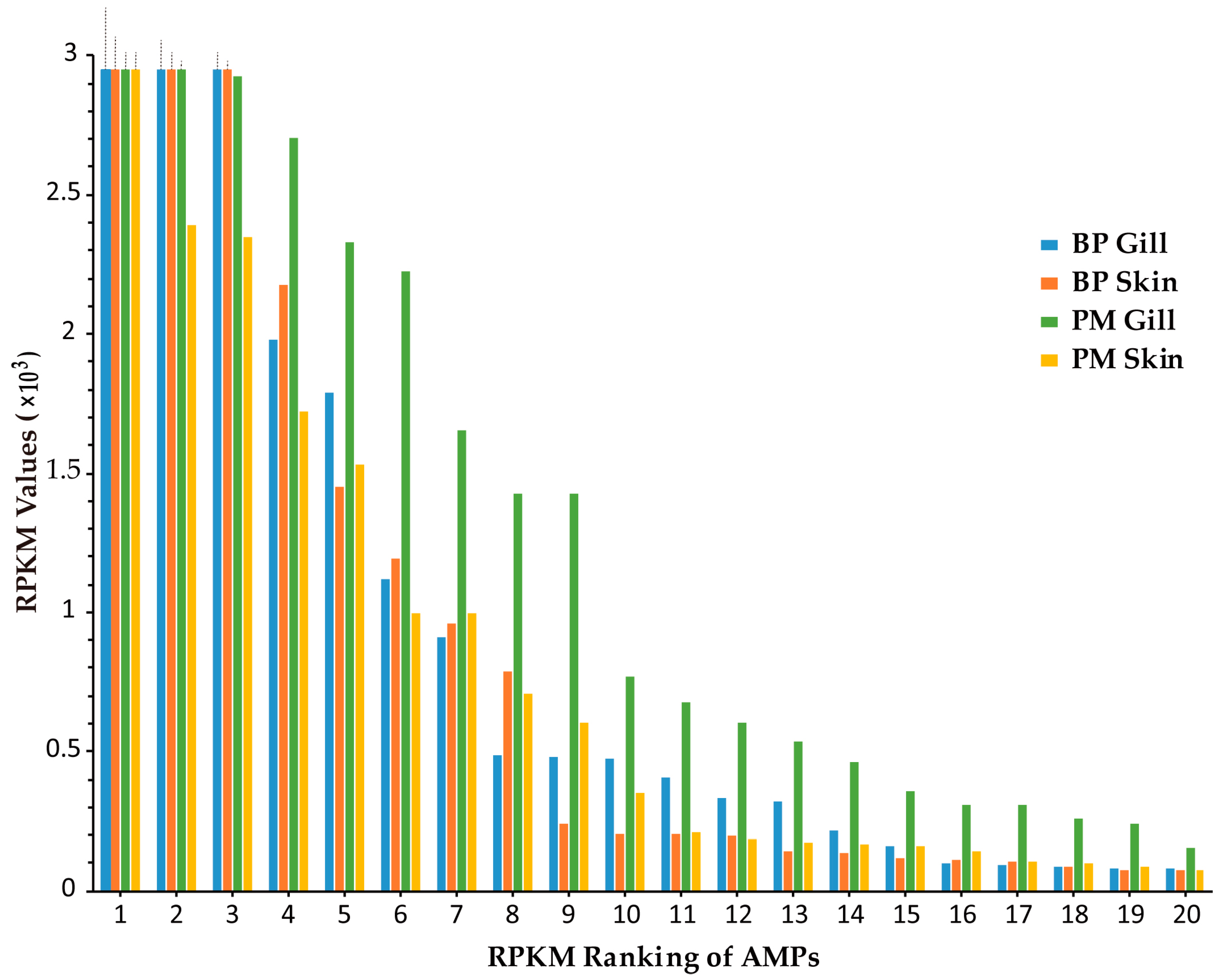
2.2. Top Ranked AMPs in Four Tissue Transcriptomes
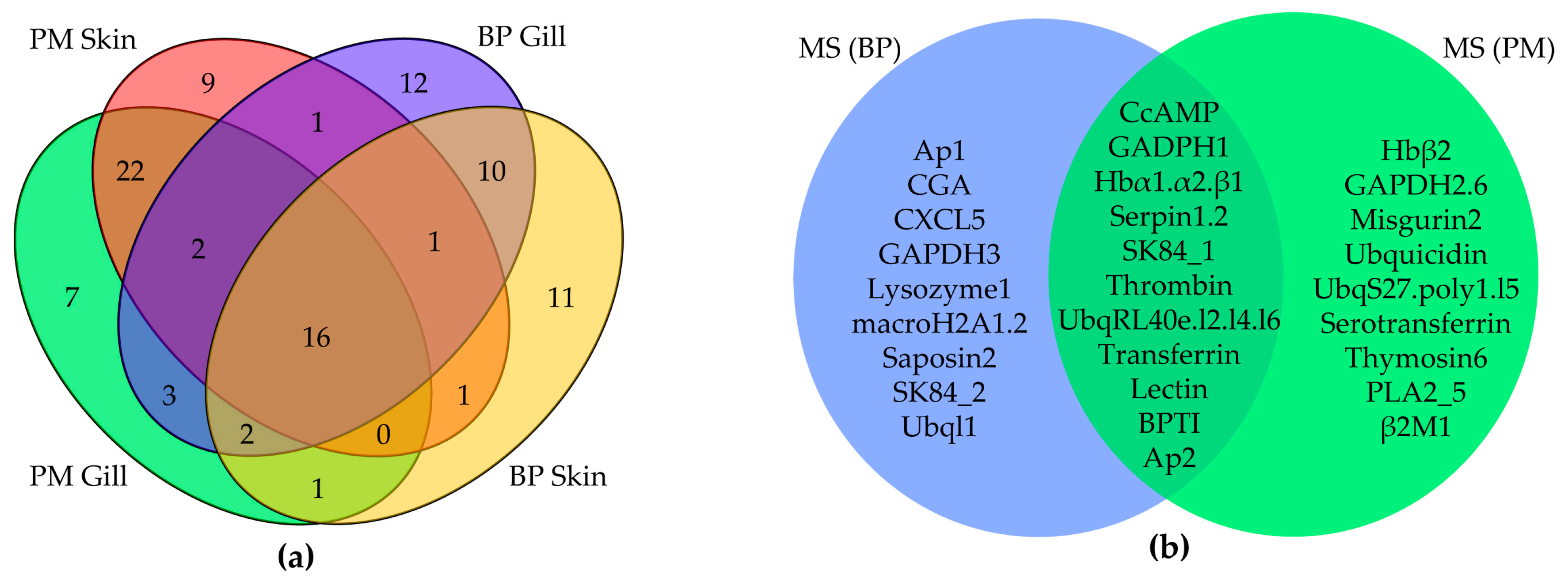
2.3. MS Validation
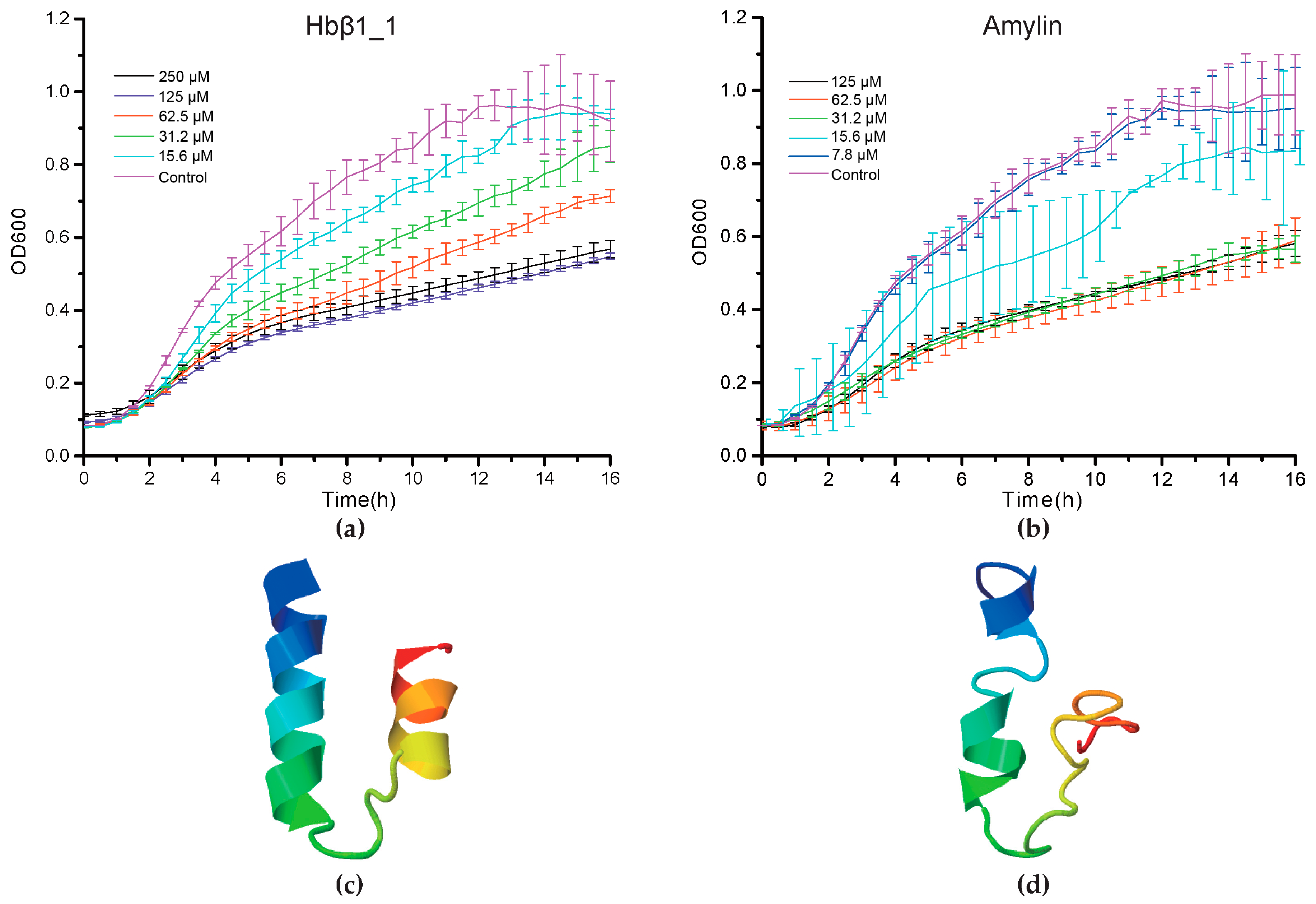
2.4. Antimicrobial Confirmation
2.5. Deep Investigations on Several Interesting AMPs
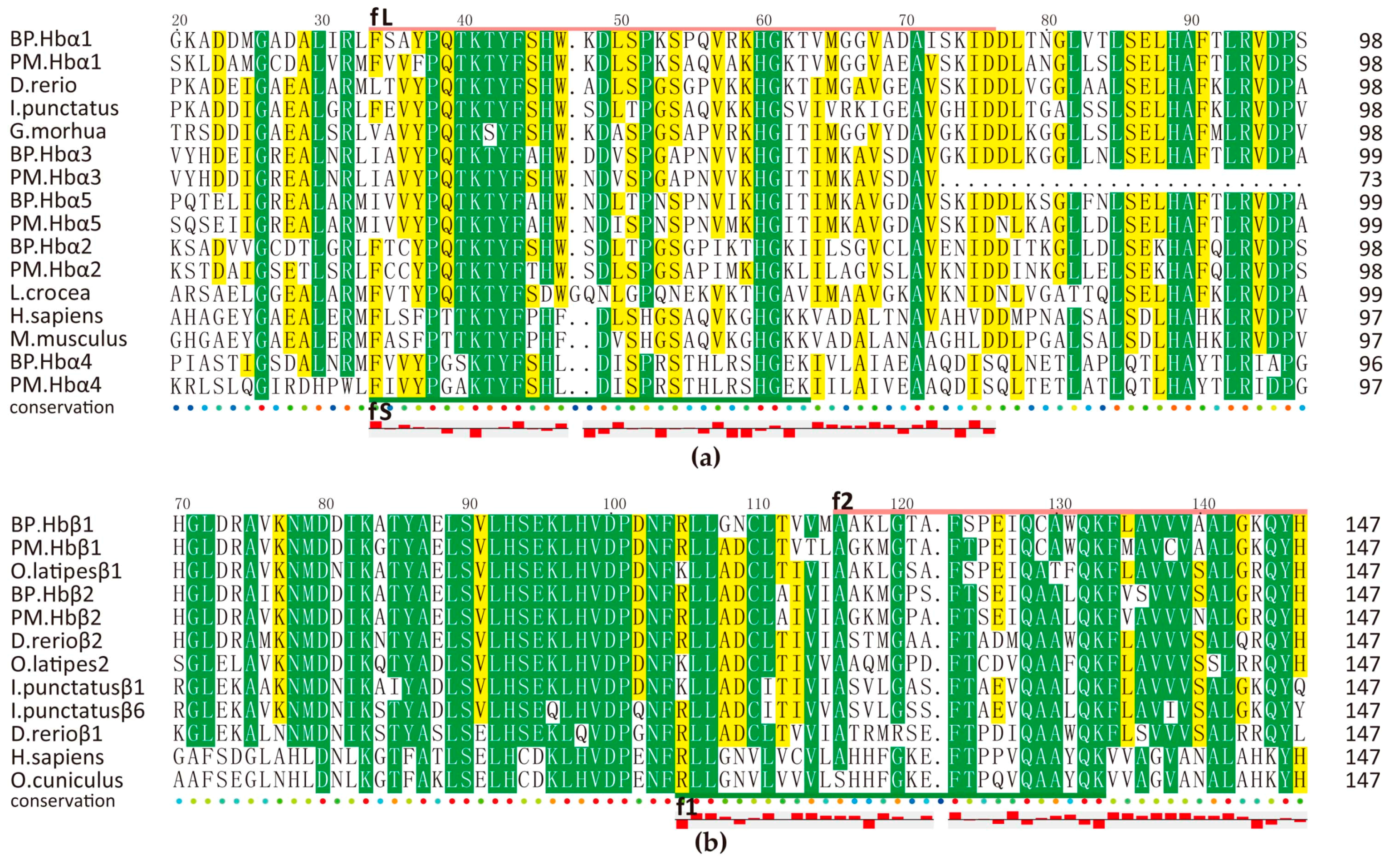
2.5.1. Hemoglobin-Derived AMPs
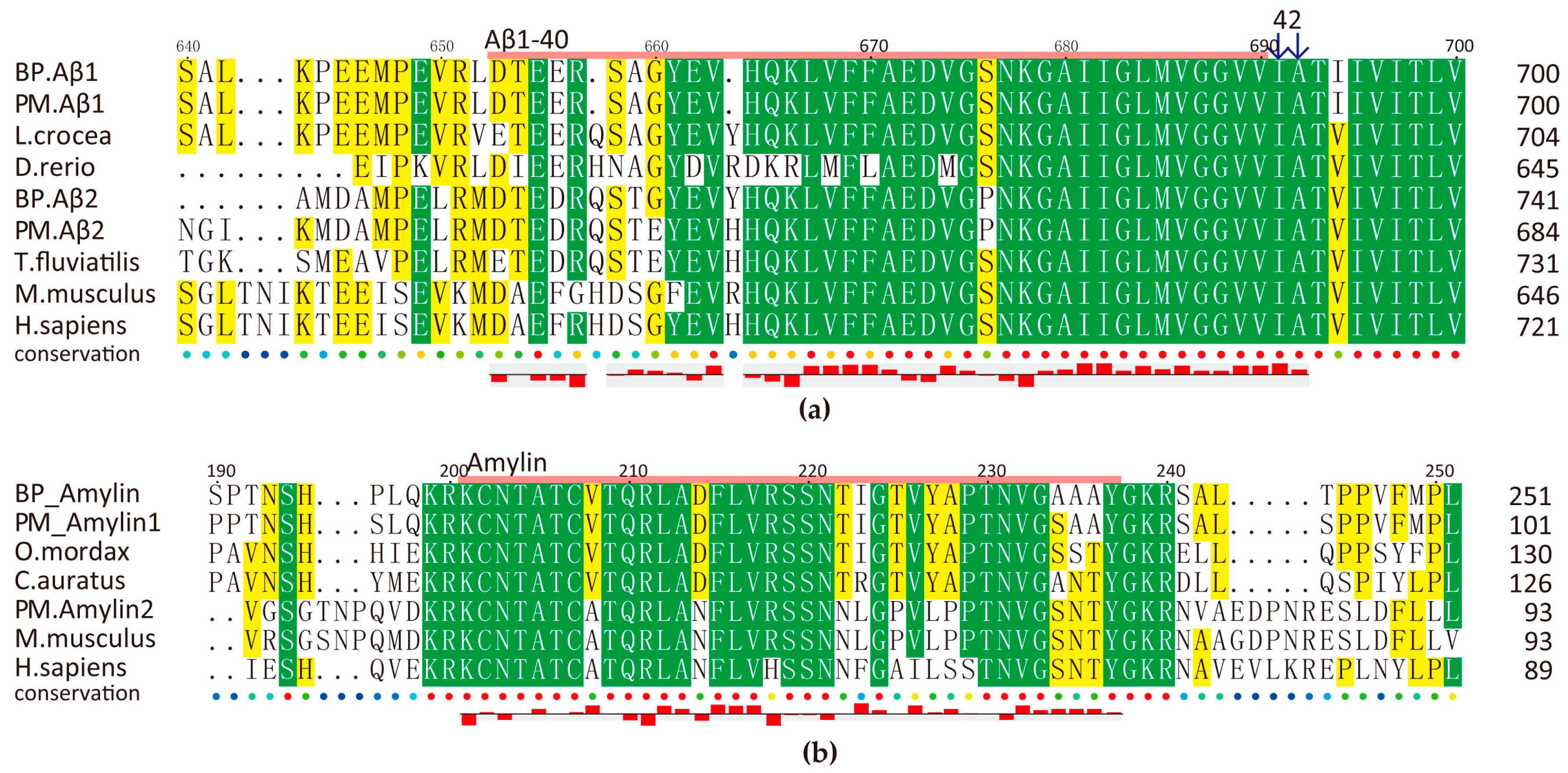
2.5.2. Amyloid AMPs
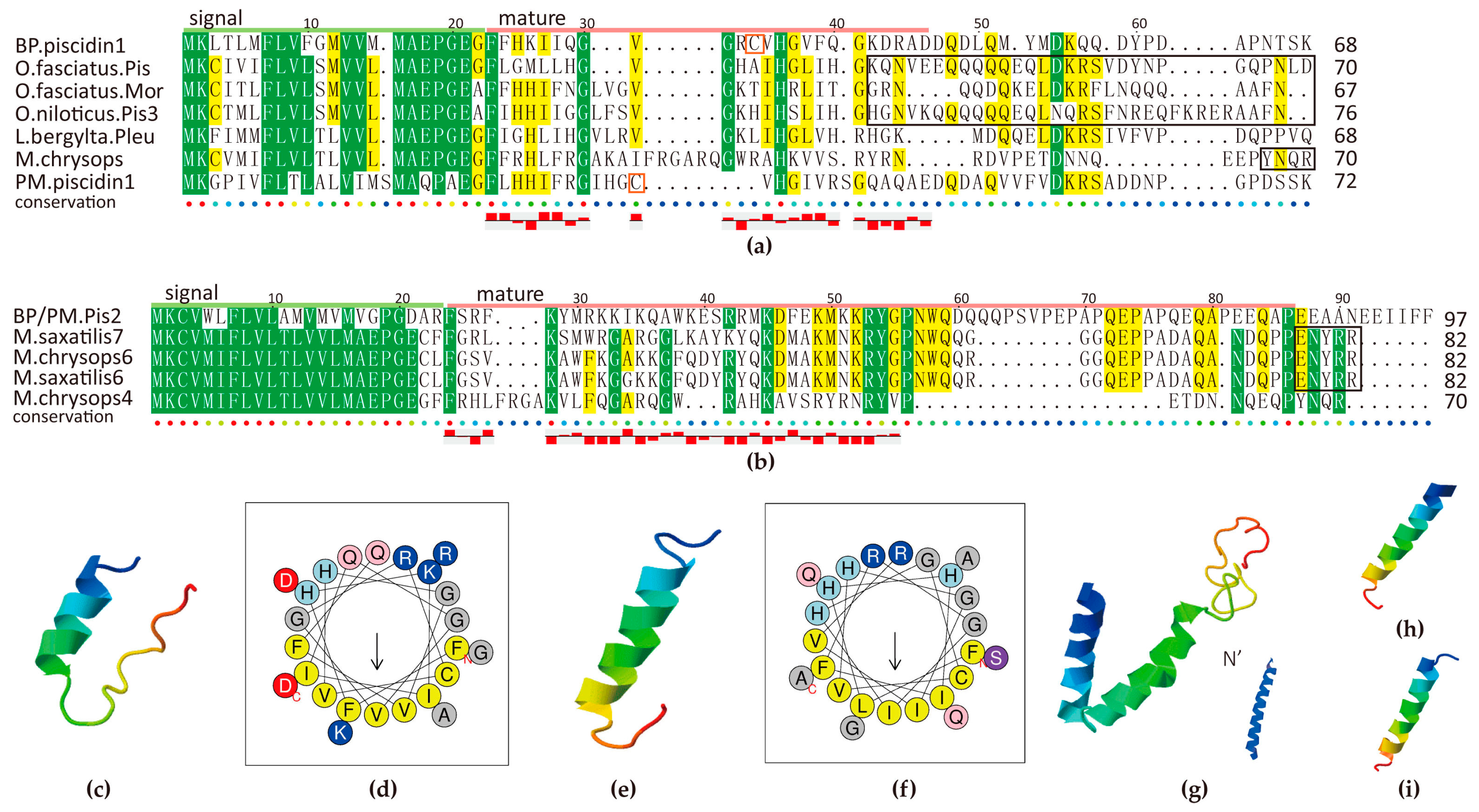
2.5.3. Piscidin Family
3. Discussion
4. Materials and Methods
4.1. Data Collection
4.2. Prediction and Identification of AMPs
4.3. MS Analysis and Protein Identification
4.4. Synthesis of Custom Peptides
4.5. Antibacterial Assays
4.6. Computational Molecular Profiling of AMPs
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Sierra, J.M.; Fusté, E.; Rabanal, F.; Vinuesa, T.; Viñas, M. An overview of antimicrobial peptides and the latest advances in their development. Expert Opin. Biol. Ther. 2017, 17, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Mansour, S.C.; Hancock, R.E. Antimicrobial peptides: An introduction. Antimicrob. Pept. Methods Protoc. 2017, 1548, 3–22. [Google Scholar]
- Patel, S.; Akhtar, N. Antimicrobial peptides (AMPs): The quintessential ‘offense and defense’ molecules are more than antimicrobials. Biomed. Pharmacother. 2017, 95, 1276–1283. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zeng, X.; Yang, Q.; Qiao, S. Antimicrobial peptides as potential alternatives to antibiotics in food animal industry. Int. J. Mol. Sci. 2016, 17, 603. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, T.; Gallo, R.L. Antimicrobial peptides: Old molecules with new ideas. J. Investig. Dermatol. 2012, 132, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Zaiou, M. Multifunctional antimicrobial peptides: Therapeutic targets in several human diseases. J. Mol. Med. 2007, 85, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Reiling, S.; Zarena, D.; Wang, G. Host defense antimicrobial peptides as antibiotics: Design and application strategies. Curr. Opin. Chem. Biol. 2017, 38, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Kindrachuk, J.; Napper, S. Structure-activity relationships of multifunctional host defence peptides. Mini Rev. Med. Chem. 2010, 10, 596–614. [Google Scholar] [CrossRef] [PubMed]
- Falanga, A.; Lombardi, L.; Franci, G.; Vitiello, M.; Iovene, M.R.; Morelli, G.; Galdiero, M.; Galdiero, S. Marine antimicrobial peptides: Nature provides templates for the design of novel compounds against pathogenic bacteria. Int. J. Mol. Sci. 2016, 17, 785. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, A.; Neundorf, I. Design and application of antimicrobial peptide conjugates. Int. J. Mol. Sci. 2016, 17, 701. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.; Naughton, L.M.; Montánchez, I.; Dobson, A.D.; Rai, D.K. Current status and future prospects of marine natural products (MNPs) as antimicrobials. Mar. Drugs 2017, 15, 272. [Google Scholar] [CrossRef] [PubMed]
- Lipkin, R.; Lazaridis, T. Computational studies of peptide-induced membrane pore formation. Philos. Trans. R. Soc. B 2017, 372. [Google Scholar] [CrossRef] [PubMed]
- Vineeth, K.T.; Sanil, G. A review of the mechanism of action of amphibian antimicrobial peptides focusing on peptide-membrane interaction and membrane curvature. Curr. Protein Pept. Sci. 2017, 18, 1263–1272. [Google Scholar]
- Travkova, O.G.; Moehwald, H.; Brezesinski, G. The interaction of antimicrobial peptides with membranes. Adv. Colloid Interface Sci. 2017, 247, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Wang, G. The importance of amino acid composition in natural AMPs: An evolutional, structural, and functional perspective. Front. Immunol. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-helical cationic antimicrobial peptides: Relationships of structure and function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Cutrona, K.J.; Kaufman, B.A.; Figueroa, D.M.; Elmore, D.E. Role of arginine and lysine in the antimicrobial mechanism of histone-derived antimicrobial peptides. FEBS Lett. 2015, 589, 3915–3920. [Google Scholar] [CrossRef] [PubMed]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Bian, C.; Zan, Q.; Xu, X.; Liu, X.; Chen, J.; Wang, J.; Qiu, Y.; Li, W.; Zhang, X.; et al. Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Agerberth, B.; Boman, A.; Andersson, M.; Jornvall, H.; Mutt, V.; Boman, H.G. Isolation of three antibacterial peptides from pig intestine: Gastric inhibitory polypeptide (7–42), diazepam-binding inhibitor (32–86) and a novel factor, peptide 3910. FEBS J. 1993, 216, 623–629. [Google Scholar] [CrossRef]
- Arenas, G.; Guzman, F.; Cardenas, C.; Mercado, L.; Marshall, S.H. A novel antifungal peptide designed from the primary structure of a natural antimicrobial peptide purified from Argopecten purpuratus hemocytes. Peptides 2009, 30, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Park, C.B.; Lee, J.H.; Park, I.Y.; Kim, M.S.; Kim, S.C. A novel antimicrobial peptide from the loach, Misgurnus anguillicaudatus. FEBS Lett. 1997, 411, 173–178. [Google Scholar] [CrossRef]
- Påhlman, L.I.; Mörgelin, M.; Kasetty, G.; Olin, A.I.; Schmidtchen, A.; Herwald, H. Antimicrobial activity of fibrinogen and fibrinogen-derived peptides—A novel link between coagulation and innate immunity. Thromb. Haemost. 2013, 109, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Papareddy, P.; Rydengård, V.; Pasupuleti, M.; Walse, B.; Morgelin, M.; Chalupka, A.; Malmsten, M.; Schmidtchen, A. Proteolysis of human thrombin generates novel host defense peptides. PLoS Pathog. 2010, 6, e1000857. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Hwang, J.S.; Lee, D.G. Identification of a novel antimicrobial peptide, scolopendin1, derived from centipede Scolopendra subspinipes mutilans and its antifungal mechanism. Insect Mol. Biol. 2014, 23, 788–799. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Lee, M.J.; Go, H.J.; Kim, G.D.; Jeong, H.D.; Nam, B.H.; Park, N.G. Purification and antimicrobial function of ubiquitin isolated from the gill of Pacific oyster, Crassostrea gigas. Mol. Immunol. 2013, 53, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, C.P.; Bogaards, S.J.; Wulferink, M.; Velders, M.P.; Welling, M.M. Synthetic peptides derived from human antimicrobial peptide ubiquicidin accumulate at sites of infections and eradicate (multi-drug resistant) Staphylococcus aureus in mice. Peptides 2006, 27, 2585–2591. [Google Scholar] [CrossRef] [PubMed]
- Hiemstra, P.S.; van den Barselaar, M.T.; Roest, M.; Nibbering, P.H.; van Furth, R. Ubiquicidin, a novel murine microbicidal protein present in the cytosolic fraction of macrophages. J. Leukoc. Biol. 1999, 66, 423–428. [Google Scholar] [PubMed]
- Coates, C.J.; Decker, H. Immunological properties of oxygen-transport proteins: Hemoglobin, hemocyanin and hemerythrin. Cell Mol. Life Sci. 2017, 74, 293–317. [Google Scholar] [CrossRef] [PubMed]
- Froidevaux, R.; Krier, F.; Nedjar-Arroume, N.; Vercaigne-Marko, D.; Kosciarz, E.; Ruckebusch, C.; Dhulster, P.; Guillochon, D. Antibacterial activity of a pepsin-derived bovine hemoglobin fragment. FEBS Lett. 2001, 491, 159–163. [Google Scholar] [CrossRef]
- Deng, L.; Pan, X.; Wang, Y.; Wang, L.; Zhou, X.E.; Li, M.; Feng, Y.; Wu, Q.; Wang, B.; Huang, N. Hemoglobin and its derived peptides may play a role in the antibacterial mechanism of the vagina. Hum. Reprod. 2009, 24, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.V.R.; Sukanya, D.; Aranha, C.; Patgaonkar, M.S.; Bhonde, G. Identification and characterization of molecules having anti-microbial activities from rabbit epididymis using proteomic approach. J. Microb. Biochem. Technol. 2011. [Google Scholar] [CrossRef]
- Ullal, A.J.; Litaker, R.W.; Noga, E.J. Antimicrobial peptides derived from hemoglobin are expressed in epithelium of channel catfish (Ictalurus punctatus, Rafinesque). Dev. Comp. Immunol. 2008, 32, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Parish, C.A.; Jiang, H.; Tokiwa, Y.; Berova, N.; Nakanishi, K.; McCabe, D.; Zuckerman, W.; Xia, M.M.; Gabay, J.E. Broad-spectrum antimicrobial activity of hemoglobin. Bioorg. Med. Chem. 2001, 9, 377–382. [Google Scholar] [CrossRef]
- Soscia, S.J.; Kirby, J.E.; Washicosky, K.J.; Tucker, S.M.; Ingelsson, M.; Hyman, B.; Burton, M.A.; Goldstein, L.E.; Duong, S.; Tanzi, R.E.; et al. The Alzheimer’s disease-associated amyloid β-protein is an antimicrobial peptide. PLoS ONE 2010, 5, e9505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Liu, Q.; Chen, J.C.; Cui, Y.X.; Zhou, B.; Chen, Y.X.; Zhao, Y.F.; Li, Y.M. Antimicrobial activity of human islet amyloid polypeptides: An insight into amyloid peptides’ connection with antimicrobial peptides. Biol. Chem. 2012, 393, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Bleackley, M.R.; Hayes, B.M.; Parisi, K.; Saiyed, T.; Traven, A.; Potter, I.D.; van der Weerden, N.L.; Anderson, M.A. Bovine pancreatic trypsin inhibitor is a new antifungal peptide that inhibits cellular magnesium uptake. Mol. Microbiol. 2014, 92, 1188–1197. [Google Scholar] [CrossRef] [PubMed]
- Salger, S.A.; Cassady, K.R.; Reading, B.J.; Noga, E.J. A diverse family of host-defense peptides (Piscidins) exhibit specialized anti-bacterial and anti-protozoal activities in fishes. PLoS ONE 2016, 11, e0159423. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.M.O.; Ruangsri, J.; Kiron, V. Atlantic cod piscidin and its diversification through positive selection. PLoS ONE 2010, 5, e9501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silphaduang, U.; Noga, E.J. Antimicrobials: Peptide antibiotics in mast cells of fish. Nature 2001, 414, 268–269. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lee, W.H.; Zhang, Y. Extremely abundant antimicrobial peptides existed in the skins of nine kinds of Chinese odorous frogs. J. Proteome Res. 2012, 11, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jiang, J.; Wu, Z.; Xie, F. Antimicrobial peptides from the skin of the Asian frog, Odorrana jingdongensis: De novo sequencing and analysis of tandem mass spectrometry data. J. Proteom. 2012, 75, 5807–5821. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Post-translational modifications of natural antimicrobial peptides and strategies for peptide engineering. Curr. Biotechnol. 2012, 1, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Henriques, S.T.; Tan, C.C.; Craik, D.J.; Clark, R.J. Structural and functional analysis of human liver-expressed antimicrobial peptide 2. Chembiochem 2010, 11, 2148–2157. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Vogel, H.J. Structural perspectives on antimicrobial chemokines. Front. Immunol. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, J.; Anboukaria, H.; Luo, Z.; Zhao, M.; Wu, H. Comparative transcriptome analyses of seven anurans reveal functions and adaptations of amphibian skin. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Pukala, T.L.; Bowie, J.H.; Maselli, V.M.; Musgrave, I.F.; Tyler, M.J. Host-defence peptides from the glandular secretions of amphibians: Structure and activity. Nat. Prod. Rep. 2006, 23, 368–393. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, C.A.; Zuloaga, R.; Poblete-Morales, M.; Vera-Tobar, T.; Mercado, L.; Avendano-Herrera, R.; Valdes, J.A.; Molina, A. Fish skeletal muscle tissue is an important focus of immune reactions during pathogen infection. Dev. Comp. Immunol. 2017, 73, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Zhang, P.; Zhang, Q.; Zhang, J. Two hepcidins from spotted scat (Scatophagus argus) possess antibacterial and antiviral functions in vitro. Fish Shellfish Immunol. 2016, 50, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Gao, Y.; Wang, R.; Xu, T. Characterization, evolution and functional analysis of the liver-expressed antimicrobial peptide 2 (LEAP-2) gene in miiuy croaker. Fish Shellfish Immunol. 2014, 41, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Meng, P.; Yang, S.; Shen, C.; Jiang, K.; Rong, M.; Lai, R. The first salamander defensin antimicrobial peptide. PLoS ONE 2013, 8, e83044. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Hepcidin and iron regulation, 10 years later. Blood 2011, 117, 4425–4433. [Google Scholar] [CrossRef] [PubMed]
- Neves, J.V.; Caldas, C.; Ramos, M.F.; Rodrigues, P.N. Hepcidin-dependent regulation of erythropoiesis during anemia in a teleost fish, Dicentrarchus labrax. PLoS ONE 2016, 11, e0153940. [Google Scholar] [CrossRef] [PubMed]
- Sheokand, N.; Malhotra, H.; Kumar, S.; Tillu, V.A.; Chauhan, A.S.; Raje, C.I.; Raje, M. Moonlighting cell-surface GAPDH recruits apotransferrin to effect iron egress from mammalian cells. J. Cell Sci. 2014, 127, 4279–4291. [Google Scholar] [CrossRef] [PubMed]
- Gudmundsson, G.H.; Agerberth, B. Neutrophil antibacterial peptides, multifunctional effector molecules in the mammalian immune system. J. Immunol. Methods 1999, 232, 45–54. [Google Scholar] [CrossRef]
- Semeraro, F.; Ammollo, C.T.; Morrissey, J.H.; Dale, G.L.; Friese, P.; Esmon, N.L.; Esmon, C.T. Extracellular histones promote thrombin generation through platelet-dependent mechanisms: Involvement of platelet TLR2 and TLR4. Blood 2011, 118, 1952–1961. [Google Scholar] [CrossRef] [PubMed]
- Bustillo, M.E.; Fischer, A.L.; LaBouyer, M.A.; Klaips, J.A.; Webb, A.C.; Elmore, D.E. Modular analysis of hipposin, a histone-derived antimicrobial peptide consisting of membrane translocating and membrane permeabilizing fragments. Biochim. Biophys. Acta 2014, 1838, 2228–2233. [Google Scholar] [CrossRef] [PubMed]
- Hoeksema, M.; van Eijk, M.; Haagsman, H.P.; Hartshorn, K.L. Histones as mediators of host defense, inflammation and thrombosis. Future Microbiol. 2016, 11, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, S.C.; Lee, J.K.; Choi, S.J.; Hahm, K.S.; Park, Y. Novel antibacterial activity of β2-microglobulin in human amniotic fluid. PLoS ONE 2012, 7, e47642. [Google Scholar] [CrossRef] [PubMed]
- Roudi, R.; Syn, N.L.; Roudbary, M. Antimicrobial peptides as biologic and immunotherapeutic agents against cancer: A comprehensive overview. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Hilpert, K.; Volkmer-Engert, R.; Walter, T.; Hancock, R.E. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 2005, 23, 1008–1012. [Google Scholar] [CrossRef] [PubMed]
- Lata, S.; Sharma, B.K.; Raghava, G.P. Analysis and prediction of antibacterial peptides. BMC Bioinform. 2007, 8. [Google Scholar] [CrossRef]
- Mura, M.; Wang, J.; Zhou, Y.; Pinna, M.; Zvelindovsky, A.V.; Dennison, S.R.; Phoenix, D.A. The effect of amidation on the behaviour of antimicrobial peptides. Eur. Biophys. J. 2016, 45, 195–207. [Google Scholar] [PubMed]
- Molchanova, N.; Hansen, P.R.; Franzyk, H. Advances in development of antimicrobial peptidomimetics as potential drugs. Molecules 2017, 22, 1430. [Google Scholar] [CrossRef]
- Li, H.X.; Lu, X.J.; Li, C.H.; Chen, J. Molecular characterization of the liver-expressed antimicrobial peptide 2 (LEAP-2) in a teleost fish, Plecoglossus altivelis: Antimicrobial activity and molecular mechanism. Mol. Immunol. 2015, 65, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Zdybicka-Barabas, A.; Mak, P.; Klys, A.; Skrzypiec, K.; Mendyk, E.; Fiolka, M.J.; Cytrynska, M. Synergistic action of Galleria mellonella anionic peptide 2 and lysozyme against Gram-negative bacteria. Biochim. Biophys. Acta 2012, 1818, 2623–2635. [Google Scholar] [CrossRef] [PubMed]
- Bjerkan, L.; Sonesson, A.; Schenck, K. Multiple functions of the new cytokine-based antimicrobial peptide thymic stromal lymphopoietin (TSLP). Pharmaceuticals 2016, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Alaiwa, M.H.A.; Reznikov, L.R.; Gansemer, N.D.; Sheets, K.A.; Horswill, A.R.; Stoltz, D.A.; Zabner, J.; Welsh, M.J. pH modulates the activity and synergism of the airway surface liquid antimicrobials beta-defensin-3 and LL-37. Proc. Natl. Acad. Sci. USA 2014, 111, 18703–18708. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Cheng, L.; Zhang, S.; Wang, L.; Hu, J. The role of natural antimicrobial peptides during infection and chronic inflammation. Antonie Van Leeuwenhoek 2017, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Sun, Y.; Shi, G.; Wang, R. Miiuy croaker hepcidin gene and comparative analyses reveal evidence for positive selection. PLoS ONE 2012, 7, e35449. [Google Scholar] [CrossRef] [PubMed]
- Tennessen, J.A. Enhanced synonymous site divergence in positively selected vertebrate antimicrobial peptide genes. J. Mol. Evol. 2005, 61, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Beitz, E. TEXshade: Shading and labeling multiple sequence alignments using LATEX2 epsilon. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Cadman, E.; Bostwick, J.R.; Eichberg, J. Determination of protein by a modified Lowry procedure in the presence of some commonly used detergents. Anal. Biochem. 1979, 96, 21–23. [Google Scholar] [CrossRef]
- Brosch, M.; Yu, L.; Hubbard, T.; Choudhary, J. Accurate and sensitive peptide identification with Mascot Percolator. J. Proteome Res. 2009, 8, 3176–3181. [Google Scholar] [CrossRef] [PubMed]
- Kent, S.B. Chemical synthesis of peptides and proteins. Annu. Rev. Biochem. 1988, 57, 957–989. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Zhang, H.; Jiang, S.; Wang, M.; Wang, L.; Song, L. Comparative study of two single CRD C-type lectins, CgCLec-4 and CgCLec-5, from pacific oyster Crassostrea gigas. Fish Shellfish Immunol. 2016, 59, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific α-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
| Species | Scaffold | Gene Sets | Brain | Gill | Liver | Muscle | Skin |
|---|---|---|---|---|---|---|---|
| BP | 98 | 239 | 131 | 174 | 118 | 107 | 103 |
| PM | 126 | 225 | 145 | 165 | 144 | 103 | 156 |
| Ranking | BP Gill | BP Skin | PM Gill | PM Skin |
|---|---|---|---|---|
| 1 | Hbα2 | Hbα2 | β2M1 | GAPDH1 |
| 2 | Hbβ1 | Hbβ1 | CCL4 | Ubq/poly1 |
| 3 | Hbα1 | Hbα1 | Hbα2 | β2M1 |
| 4 | Piscidin2 | GAPDH1 | CXCL8 | Ubq/RPL40e |
| 5 | Ubiquicidin | Ubiquicidin | Ubq/poly1 | Hbα2 |
| 6 | Ubq/RPL40e | Ubq/RPL40e | Ubq/RPL40e | Hbβ1 |
| 7 | GAPDH3 | CCL1 | Hbβ1 | Ubq/RPS27a |
| 8 | DWD2 | GAPDH3 | CCL2_2 | GAPDH6 |
| 9 | Thrombin8 | DWD2 | Ubq/RPS27a | Misgurin2 |
| 10 | Defensinα2 | CCL3 | Hbα1 | Hbα1 |
| 11 | CCL1 | Saposin2 | GAPDH1 | SK84_2 |
| 12 | Piscidin1 | H2A5 | SK84_2 | SK84_1 |
| 13 | Saposin2 | CCL2 | CCL2 | Serotransferrin |
| 14 | CXCL7 | BPTI6 | Thrombin40 | CXCL8 |
| 15 | CCL3 | Hepcidin2 | CXCL7 | Saposin2 |
| 16 | GAPDH1 | SWD3 | GAPDH6 | Ubiquicidin |
| 17 | CCL2 | RNase6 | CCL3 | BPTI18 |
| 18 | Defensinα1 | Glyrichin | Saposin2 | BPTI24 |
| 19 | SWD3 | CXCL7 | Thrombin9 | Ap |
| 20 | Glyrichin | CCL4 | H2A1 | CcAMP |
| Sample | Total/Identified Spectra | AMP Spectra | Identified Peptides/Proteins | AMP Peptides/Proteins |
|---|---|---|---|---|
| BP Skin | 168,975/65,778 | 3843 | 10,985/1292 | 308/45 |
| BP Muscle | 137,162/49,488 | 1313 | 5140/435 | 124/17 |
| PM Skin | 166,870/68,187 | 4013 | 9898/1442 | 270/47 |
| PM Muscle | 144,915/52,062 | 1417 | 4787/577 | 155/22 |
| Tissue | Hbβ1 | Hbβ2 | Hbα1 | Hbα2 | Hbα3 | Hbα4 | Hbα5 |
|---|---|---|---|---|---|---|---|
| BP Brain | 3691.38 | 2.72 | 2909.06 | 16,212.60 | 0.20 | 4.82 | 0.44 |
| PM Brain | 128.06 | 0 | 63.62 | 230.17 | 0 | 0 | 0 |
| BP Gill | 12,092.00 | 7.21 | 6047.51 | 41,419.70 | 0 | 10.05 | 0.04 |
| PM Gill | 1652.96 | 0 | 768.13 | 2928.19 | 0 | 0.06 | 0 |
| BP Liver | 1767.70 | 0.19 | 440.72 | 5545.23 | 0 | 2.99 | 0 |
| PM Liver | 4448.83 | 0.24 | 1422.59 | 8908.93 | 0 | 0.09 | 0 |
| BP Muscle | 4927.71 | 0.65 | 2460.18 | 14,000.1 | 0 | 1.75 | 0.06 |
| PM Muscle | 919.64 | 0 | 339.07 | 1678.62 | 0 | 0.08 | 0 |
| BP Skin | 7408.15 | 4.75 | 4211.22 | 17,282.30 | 0.57 | 5.31 | 0 |
| PM Skin | 996.73 | 0 | 354.46 | 1529.95 | 0 | 0.10 | 0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, Y.; You, X.; Bian, C.; Chen, S.; Lv, Z.; Qiu, L.; Shi, Q. High-Throughput Identification of Antimicrobial Peptides from Amphibious Mudskippers. Mar. Drugs 2017, 15, 364. https://doi.org/10.3390/md15110364
Yi Y, You X, Bian C, Chen S, Lv Z, Qiu L, Shi Q. High-Throughput Identification of Antimicrobial Peptides from Amphibious Mudskippers. Marine Drugs. 2017; 15(11):364. https://doi.org/10.3390/md15110364
Chicago/Turabian StyleYi, Yunhai, Xinxin You, Chao Bian, Shixi Chen, Zhao Lv, Limei Qiu, and Qiong Shi. 2017. "High-Throughput Identification of Antimicrobial Peptides from Amphibious Mudskippers" Marine Drugs 15, no. 11: 364. https://doi.org/10.3390/md15110364





