Inhibitors of Serine Proteases from a Microcystis sp. Bloom Material Collected from Timurim Reservoir, Israel
Abstract
:1. Introduction
2. Results and Discussion
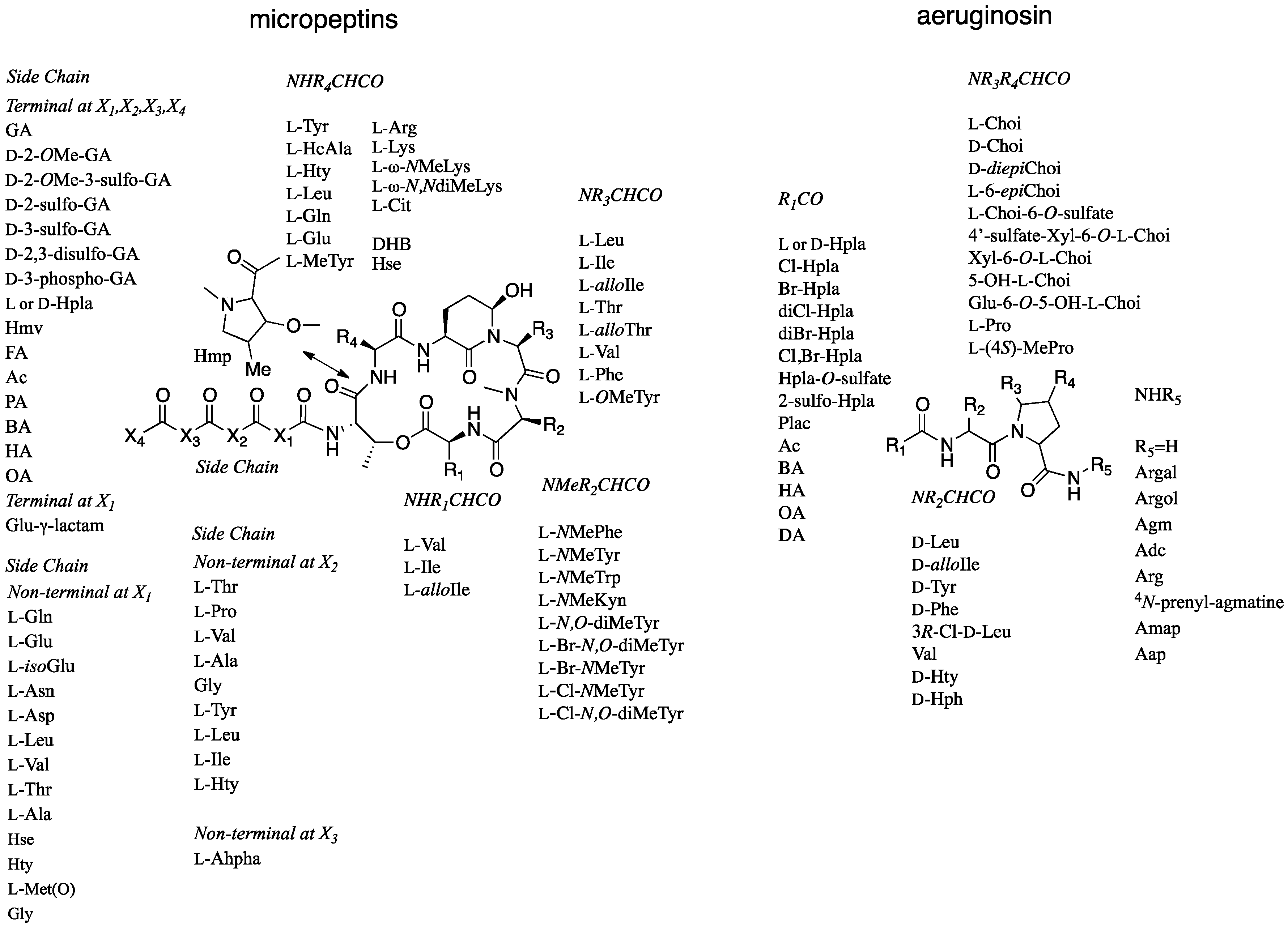
2.1. Structural Elucidation of Micropeptin TR1058
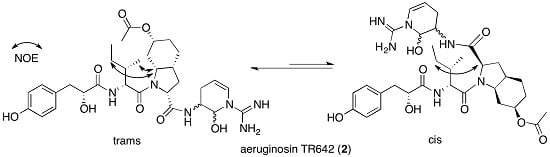
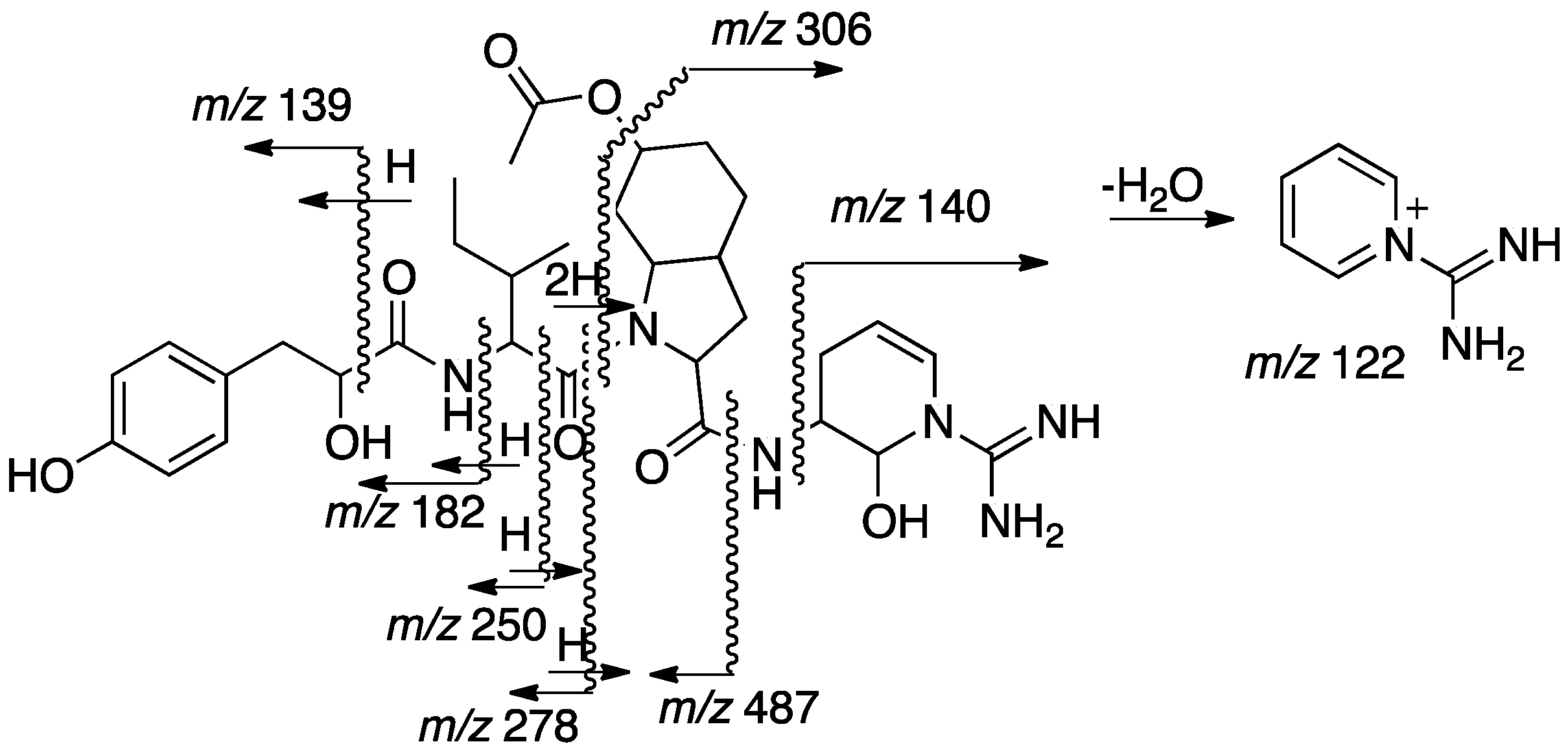
2.2. Structural Elucidation of Aeruginosin TR642
2.3. Biological Activity Assessment
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Biological Material
3.3. Isolation Procedure
3.4. Determination of the Absolute Configuration of the Amino Acids by Marfey’s Method
3.5. Determination of the Absolute Configuration of Hydroxy Phenyl Lactic Acid (Hpla)
3.6. Protease Inhibition Assays
3.6.1. Trypsin
3.6.2. Thrombin
3.6.3. Chymotrypsin
3.6.4. Elastase
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gademann, K.; Portmann, C. Secondary metabolites from cyanobacteria: Complex structures and powerful bioactivities. Curr. Org. Chem. 2008, 12, 326–341. [Google Scholar] [CrossRef]
- Pearson, L.; Mihali, T.; Moffitt, M.; Kellmann, R.; Neilen, B. On the chemistry, toxicology and genetics of the cyanobacterial toxins, microcystins, nodularin, saxitoxin and cylindrospermopsin. Mar. Drugs 2010, 8, 1650–1680. [Google Scholar] [CrossRef] [PubMed]
- Teta, R.; Della Sala, G.; Glukhov, E.; Gerwick, L.; Gerwick, W.H.; Mangoni, A.; Constantino, V. Combined LC-MS/MS and molecular netwarking approch reveals new cyanotoxins from the 2014 cyanobacterial bloom in Green Lake, Seattle. Environ. Sci. Technol. 2015, 49, 14301–14310. [Google Scholar] [CrossRef] [PubMed]
- Bittencourt-Oliveira, M.C.; Herman, T.C.; Cordeiro-Araujo, M.K.; Macedo-Silva, I.; Dias, C.T.; Sasaki, F.F.C.; Moura, A.N. Phytotoxicity associated to microcystins: A review. Braz. J. Biol. 2014, 74, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Oberer, L.; Ino, T.; Konig, W.A.; Busch, M.; Weckesser, J. Cyanopeptolins, new depsipeptides from the cyanobacterium Microcystis sp. PCC 7806. J. Antibiot. 1993, 46, 1550–1556. [Google Scholar] [CrossRef] [PubMed]
- Ersmark, K.; Del Valle, J.R.; Hanessian, S. Chemistry and biology of the aeruginosin family of serine protease inhibitors. Angew. Chem. Int. Ed. 2008, 47, 1202–1223. [Google Scholar] [CrossRef] [PubMed]
- Elkobi-Peer, S.; Carmeli, S. New prenylated aeruginosin, microphycin, anabaenopeptin and micropeptin analogues from a Microcystis bloom material collected in Kibbutz Kfar Blum, Israel. Mar. Drugs 2015, 13, 2347–2375. [Google Scholar] [CrossRef] [PubMed]
- Vegman, R.; Carmeli, S. Three aeruginosins and a microviridin from a bloom assembly of Microcystis spp. collected from a fishpond near Kibbutz Lehavot HaBashan, Israel. Tetrahedron 2014, 70, 6817–6824. [Google Scholar] [CrossRef]
- Gesner-Apter, S.; Carmeli, S. Protease inhibitors from a water bloom of the cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 2009, 72, 1429–1436. [Google Scholar] [CrossRef] [PubMed]
- Welker, M.; Marsalek, B.; Sejnhova, L.; von Dohren, H. Cyanobacterial peptides-nature’s own combinatorial biosynthesis. Peptides 2006, 27, 2090–2103. [Google Scholar] [CrossRef] [PubMed]
- Carmeli, S.; Tel-Aviv University, Tel Aviv, Israel. Unpublished work. 2017.
- Vegman, R.; Carmeli, S. Eight micropeptins from a Microcystis spp. bloom collected from a fishpond near Kibbutz Lehavot HaBashan, Israel. Tetrahedron 2013, 69, 10108–10115. [Google Scholar] [CrossRef]
- Marfey, P. Determination of d-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1984, 49, 591–596. [Google Scholar] [CrossRef]
- Lifshits, M.; Carmeli, S. Metabolites of Microcystis aeruginosa Bloom Material from Lake Kinneret, Israel. J. Nat. Prod. 2012, 75, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Bowden, K.; Heilbron, I.M.; Jones, E.R.H.; Weedon, B.C.L. Researches on acetylenic compounds. Part I. The preparation of acetylenic ketones by oxidation of acetylenic carbinols and glycols. J. Chem. Soc. 1946, 39–45. [Google Scholar] [CrossRef]
- Raveh, A.; Carmeli, S. Two novel biological active modified peptides from the cyanobacterium Microcystis sp. Phytochem. Lett. 2009, 2, 10–14. [Google Scholar] [CrossRef]
- Adiv, S.; Carmeli, S. Protease Inhibitors from Microcystis aeruginosa Bloom Material Collected from the Dalton Reservoir, Israel. J. Nat. Prod. 2013, 76, 2307–2315. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Okita, Y.; Matsuda, H.; Okino, T.; Murakami, M. Aeruginosins, protease inhibitors from the cyanobacterium Microcystis aeruginosa. Tetrahedron 1999, 55, 10971–10988. [Google Scholar] [CrossRef]
- Matsuda, H.; Okino, T.; Murakami, M.; Yamaguchi, K. Aeruginosins 102-A and B, new thrombin inhibitors from the cyanobacterium Microcystis viridis (NIES-102). Tetrahedron 1996, 52, 14501–14506. [Google Scholar] [CrossRef]
- Kodani, S.; Ishida, K.; Murakami, M. Aeruginosin 103-A, a thrombin inhibitor from the cyanobacterium microcystis viridis. J. Nat. Prod. 1998, 61, 1046–1048. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Welker, M.; Christiansen, G.; Cadel-Six, S.; Couchier, C.; Dittmann, E.; Hertweck, C.; Tanddeau de Marsac, N. Plasticity and evolution of aeruginosin biosynthesis in cyanobacteria. Appl. Environ. Microbiol. 2009, 75, 2017–2029. [Google Scholar] [CrossRef] [PubMed]
- Fujii, K.; Ikai, Y.; Mayumi, T.; Oka, H.; Suzuki, M.; Harada, K.A. Nonempirical method using LC/MS for determination of the absolute configuration of constituent amino acids in a peptide: Elucidation of limitations of marfey’s method and of its separation mechanism. Anal. Chem. 1997, 69, 3346–3352. [Google Scholar] [CrossRef]
- Blom, J.F.; Bister, B.; Bischoff, D.; Nicholson, G.; Jung, G.; Sussmuth, R.D.; Juttner, F. Oscillapeptin J, a new grazer toxin of the freshwater Cyanobacterium Planktothrix rubescens. J. Nat. Prod. 2003, 66, 431–434. [Google Scholar] [CrossRef] [PubMed]
- Okino, T.; Qi, S.; Matsuda, H.; Murakami, M.; Yamaguchi, K. Nostopeptins A and B, elastase inhibitors from the cyanobacterium Nostoc minutum. J. Nat. Prod. 1997, 60, 158–161. [Google Scholar] [CrossRef]
- Nishizawa, T.; Ueda, A.; Nakano, T.; Nishizawa, A.; Miura, T.; Asayama, M.; Fujii, K.; Harada, K.-I.; Shirai, M. Characterization of the locus of genes encoding enzymes producing heptadepsipetide micropeptin in the unicellular cyanobacterium Microcystis. J. Biochem. 2011, 149, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.-L.; Singh, R.; Alkhala, L.M.; Kuatsjah, E.; He, H.-Y.; Eltis, L.D.; Ryan, K.S. A pyridoxal phosphate-dependent enzyme that oxidizes an unactivated carbon-carbon bond. Nat. Chem. Biol. 2016, 12, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Christiansen, G.; Yoshida, W.Y.; Kurmayer, R.; Welker, M.; Valls, N.; Bonjoch, J.; Hertweck, C.; Borner, T.; Hemscheidt, T.; Dittmann, E. Biosynthesis and structure of aeruginoside 126A and 126B, cyanobacterial peptide glycosides bearing a 2-carboxy-6-hydroxyoctahydroindole moiety. Chem. Biol. 2007, 14, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Thorskov Bladt, T.; Kalifa-Aviv, S.; Ostenfeld Larsen, T.; Carmeli, S. Micropeptins from Microcystis sp. collected in Kabul Reservoir, Israel. Tetrahedron 2014, 70, 936–943. [Google Scholar] [CrossRef]
- Anas, A.R.J.; Mori, A.; Tone, M.; Naruse, C.; Nakajima, A.; Asukabe, H.; Takaya, Y.; Imanishi, S.Y.; Nishizawa, M.S.; Harada, K.-I. FVIIa-sTF and thrombin inhibitory activities of compounds isolated from microcystis aeruginosa K-139. Mar. Drugs 2017, 15, 275. [Google Scholar] [CrossRef] [PubMed]
- Sebaugh, J.L. Guidelines for accurate EC50/IC50 estimation. Pharm. Stat. 2011, 10, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Chlipala, G.E.; Mo, S.; Orjala, J. Chemodiversity in freshwater and terrestrial cyanobacteria–a source for drug discovery. Curr. Drugs Target. 2001, 12, 1654–1673. [Google Scholar] [CrossRef]
- Mi, Y.; Zhang, J.; He, S.; Yan, X. New peptides isolated from marine cyanobacteria, an overview over the past decade. Mar. Drugs 2017, 15, 132. [Google Scholar] [CrossRef] [PubMed]

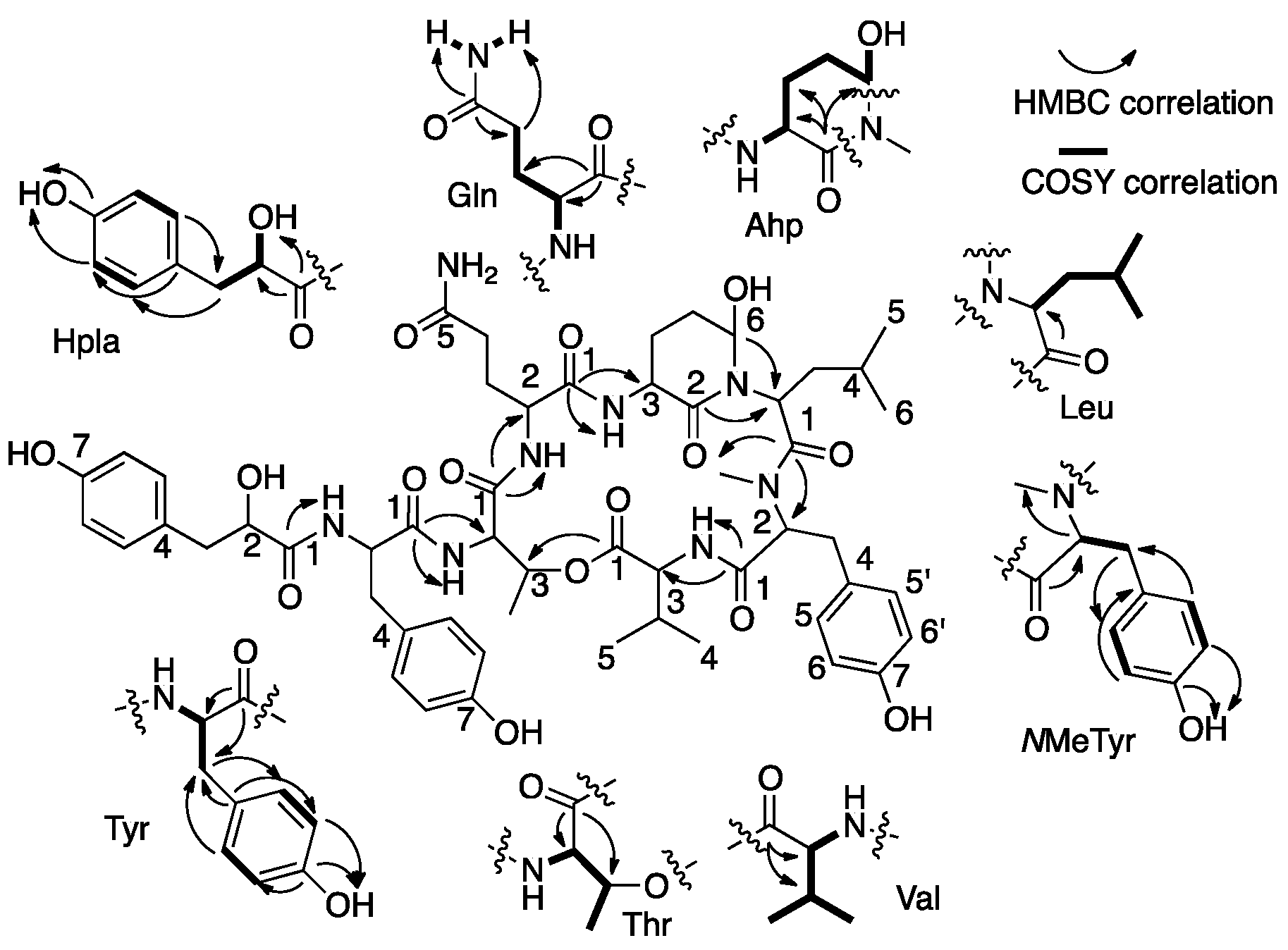
| Position | δC, Type b | δH, mult., J (Hz) | LR CH Correlations c | NOEs d |
|---|---|---|---|---|
| Val | - | - | - | - |
| 1 | 172.5, C | - | - | - |
| 2 | 56.2, CH | 4.72, dd (9.0, 5.0) | Val-1,3,4,5, NMeTyr-1 | Val-3,4,5, Ahp-OH |
| 3 | 9.30, CH | 0.27, m | Val-1,2,4,5 | Val-2,4,5,NH, Ahp-OH |
| 4 | 17.5, CH3 | 73.0, d (6.8) | Val-2,3,5 | Val-2,3,NH, Ahp-OH, NMeTyr-NMe |
| 5 | 19.5, CH3 | 0.85, d (6.8) | Val-2,3,4 | Val-2,3, NMeTyr-NMe |
| NH | - | 7.54, d (9.4) | NMeTyr-1 | Val-2,3,4, NMeTyr-2,6,6′, Ahp-OH |
| NMeTyr | - | - | - | - |
| 1 | 169.6, C | - | - | - |
| 2 | 61.1, CH | 9.41, m | NMeTyr-1,3,4,NMe, Leu-1 | NMeTyr-3a,3b,5, 5′, Val-NH, Leu-2, Ahp-OH |
| 3a | 33.1, | 2.68, d (13.9, 12.1) | NMeTyr-1,2,5,5′ | NMeTyr-2,3a,5,5′ |
| 3b | CH2 | 3.10, brd (13.9) | NMeTyr-2,5,5′ | NMeTyr-2,3b,5,5′ |
| 4 | 127.5, C | - | - | - |
| 5,5′ | 130.2, CH | 6.89, d (8.3) | NMeTyr-3,5′,5,6,6′,7 | NMeTyr-2,3a,3b,6,6′ Leu-5 |
| 6,6′ | 115.5, CH | 6.62, d (8.3) | NMeTyr-4,5,5′,6′,6,7 | NMeTyr-5,5′,OH, Leu-5 |
| 7 | 156.2, C | - | - | - |
| OH | - | 9.20 s | NMeTyr-5,5′,6,6′,7 | NMeTyr-6,6′, Leu-5 |
| NMe | 30.6, CH3 | 2.70 s | NMeTyr-2, Leu-1 | Val-4,5, Ahp-OH |
| Leu | - | - | - | - |
| 1 | 171.0, C | - | - | - |
| 2 | 47.9, CH | 4.60, dd (10.5, 3.4) | Leu-1,3,4, Ahp-2,6 | Leu-3a,4,5,6, NMeTyr-2 |
| 3a | 38.7, | 0.42, dt (10.1, 2.8) | Leu-2 | Leu-2,3b, Ahp-6 |
| 3b | CH2 | 1.54, dt (10.8, 3.0) | Leu-2 | Leu-3a, Ahp-6 |
| 4 | 23.8, CH | 0.97, m | Leu-5,6 | Leu-2, Ahp-3 |
| 5 | 22.3, CH3 | 0.49, d (6.4) | Leu-3,4,6 | Leu-2, NMeTyr-3,5,5′,6,6′,OH |
| 6 | 24.1, CH3 | 0.68, d (6.5) | Leu-2,3,4,5 | Leu-2, Ahp-3 |
| Ahp | - | - | - | - |
| 2 | 169.3, C | - | - | - |
| 3 | 49.2, CH | 4.37, ddd (11.5, 9.2, 7.0) | Ahp-2,4, Gln-1 | Ahp-5,NH, Leu-4,6 |
| 4a | 22.0, | 1.71, m | Ahp-2,5,6 | Ahp-4b |
| 4b | CH2 | 2.54, m | Ahp-3,5 | Ahp-4a |
| 5a | 30.0, | 1.71, m | Ahp-3 | Ahp-3,6,OH |
| 5b | CH2 | 1.71, m | Ahp-3 | Ahp-3,6,OH |
| 6 | 73.6, CH | 4.89, brs | Ahp-3,4 | Ahp-5,OH, Leu-3b |
| NH | - | 7.34, d (9.2) | Ahp-3, Gln-1 | Ahp-3, Gln-2 |
| OH | - | 6.02, d (2.8) | - | Ahp-5,6, NMeTyr -2, NMe, Val-2,3, 4,NH, Leu-2 |
| Gln | - | - | - | - |
| 1 | 170.2, C | - | - | - |
| 2 | 52.3, CH | 4.30, ddd (12.3, 8.5, 4.0) | Gln-1,3,4, Thr-1 | Gln-3a,3b,4a,4b, NH, Ahp-NH |
| 3a | 27.0, CH2 | 1.66, m | Gln-1,2,4,5 | Gln-2,3b,NH |
| 3b | - | 2.18, m | Gln-1,2,4 | Gln-2,3a,NH |
| 4a | 31.9, CH2 | 2.02, m | Gln-2,3,5 | Gln-2,4b,NH2(b) |
| 4b | 2.10, m | Gln-2,3,5 | Gln-2,4a,NH2(b) | |
| 5 | 173.9, C | - | - | - |
| NH | - | 8.50, d (8.5) | Gln-2,NH, Thr-1,2 | Gln-2,3a,3b, Thr-2,3,4 |
| NH2(a) | - | 6.74, s | Gln-4,5 | Gln-4a,4b |
| NH2(b) | 7.20, s | Gln-5 | ||
| Thr | - | - | - | - |
| 1 | 169.4, C | - | - | - |
| 2 | 55.0, CH | 4.67, brd (9.4) | Thr-1,3,4, Tyr-1 | Thr-3,4, Gln-NH |
| 3 | 72.1, CH | 5.50, brq (6.5) | Thr-1,4 Val-1 | Thr-2,4, Gln-NH |
| 4 | 18.0, CH3 | 1.20, d (6.4) | Thr-1,2,3 | Thr-2,3,NH |
| NH | - | 8.25, d (9.4) | Tyr-1 | Thr-4, Tyr-2,3a, 3b |
| Tyr | - | - | - | - |
| 1 | 171.9, C | - | - | - |
| 2 | 53.3, CH | 4.71, m | Tyr-1,3,4, Hpla-1 | Tyr-3a,3b,5,5′, NH, Thr-NH |
| 3a | 37.0, CH2 | 2.80, dd (13.9, 8.9) | Tyr-1,2,4,5,5′ | Tyr-2,3b,5,5′,NH |
| 3b | 2.96, dd (13.9, 3.3) | Tyr-1,2,4,5,5′ | Tyr-2,3a,5,5′,NH | |
| 4 | 127.4, C | - | - | - |
| 5,5′ | 130.5, CH | 6.98, d (8.3) | Tyr-3,5′,5,6,6′,7 | Tyr-2,3a,3b,6,6′ |
| 6,6′ | 115.0, CH | 6.60, d (8.3) | Tyr-4,5,5′,6,6′,7 | Tyr-5,5′,OH |
| 7 | 156.0, C | - | - | - |
| OH | - | 9.11, s | Tyr-5,5′,6,6′,7 | Tyr-6,6′ |
| NH | - | 7.70, d (8.2) | Tyr-1,2, Hpla-1 | Tyr-2,3a,3b, Hpla-2,OH |
| Hpla | - | - | - | - |
| 1 | 173.3, C | - | - | - |
| 2 | 72.5, CH | 3.94, ddd (9.0, 5.8, 2.4) | Hpla-1,3,4 | Hpla-3a,3b,5,5′,2-OH |
| 3a | 39.7, CH2 | 2.45, dd (13.9, 9.0) | Hpla-2,4,5,5′ | Hpla-2,3b |
| 3b | 2.78, dd (13.9, 2.4) | Hpla-2,4,5,5′ | Hpla-2,3a | |
| 4 | 128.9, C | - | - | - |
| 5,5′ | 130.4, CH | 6.93, d (8.4) | Hpla-3,5′,5,6,6′,7 | Hpla-2,6,6′ |
| 6,6′ | 114.9, CH | 6.60, d (8.4) | Hpla-4,5,5′,6′,6,7 | Hpla-5,5′,OH |
| 7 | 155.7, C | - | - | - |
| 2-OH | - | 5.37, d (5.8) | Hpla-1,2,3 | Hpla-2, Tyr-NH |
| 7-OH | - | 9.07, s | Hpla-5,5′,6,6′,7 | Hpla-6,6′ |
| Position | δC, Type b | δH, mult. | LR CH Corr. c | NOE Corr. d |
|---|---|---|---|---|
| Hpla | - | - | - | - |
| 1 | 172.92–173.40, C | - | - | - |
| 2 | 72.28, 72.36, CH | 4.06, m | Hpla-4, | Hpla-3a,3b,5, Ile-2,NH |
| 3a | 39.5, CH2 | 2.60–2.62, m | Hpla-1,2,4,5 | Hpla-2,3b,5,2-OH, Ile-NH |
| 3b | 2.84–2.88, m | Hpla-1,2,4,5 | Hpla-2,3a,5 | |
| 4 | 128.21–128.47 C | - | - | - |
| 5,5′ | 130.50–130.64, CH | 6.98–7.00, d (8.6) | Hpla-2,3,5′,5 | Hpla-2,3a,3b,6,6′ |
| 6,6′ | 114.86, 114.90, CH | 6.61–6.63, d (8.6) | Hpla-4,5,5′,6′,6 | Hpla-5,7-OH |
| 7 | 155.88, C | - | - | - |
| 2-OH | - | 5.48–5.68, m | Hpla-1,2,3 | - |
| 7-OH | - | 9.11, s | Hpla-6,6′,7 | Hpla-6 |
| Ile | - | - | - | - |
| 1 | 169.25–169.74, C | - | - | - |
| 2 | 53.12–53.60, CH | 4.02–4.40, m | Hpla-1, Ile-1,3,4,6 | Ile-3,4a,4b,5,6,NH, Choi-2, Hpla-2 |
| 3 | 34.96–37.42, CH | 1.59–1.64, m | - | Ile-2,4a,4b,6,NH |
| 4a | 26.63–26.75, CH2 | 1.10–1.19, m | Ile-3,5,6 | Ile-3,4b,5 |
| 4b | 0.82–0.93, m | Ile-2,5,6 | Ile-3,4a,5 | |
| 5 | 11.68–12.14, CH3 | 0.71, m–0.81, t (6.8) | Ile-3,4 | Ile-2,4a,4b, Hpla-5,6 |
| 6 | 13.52–14.29, CH3 | 0.66, brd (5.8)–0.73, d (6.4) | Ile-2,3,4 | Ile-2,3,NH |
| NH | - | 7.37–7.50, m | Hpla-1, Ile-2 | Ile-2,3,6, Hpla-2,3a |
| Choi | - | - | - | - |
| 1 | 171.3–172.22, C | - | - | - |
| 2 | 58.42–59.33, CH | 4.18, d (9.0)–4.71, m | Choi-1,3,3a,7a, Ile-1 | Ile-2, Choi-3ax,3eq |
| 3eq | 30.65–33.61, CH2 | 2.21–2.27, m | Choi-1 | Choi-2,3ax |
| 3ax | 1.34–1.68, m | Choi-1,7a | Choi-2,3eq | |
| 3a | 34.60–31.99, CH | 2.31–2.51, m | Choi-2,7a | - |
| 4ax | 21.78–22.84, CH2 | 1.44–2.03, m | Choi-5 | Choi-6 |
| 4eq | 2.15–2.49, m | Choi-7a | ||
| 5ax | 26.01, | 1.44, m | Choi-4,6 | Choi-7ax |
| 5eq | 25.38, CH2 | 1.63–1.68, m | Choi-6 | Choi-6, |
| 6 | 70.65, CH | 4.55, m, 4.49, dt (3.4,11.2) | Ac-1 | Choi-4ax,5eq,7eq,7a |
| 7ax | 31.66, | 0.97, m, 1.42, m | Choi-6,7a | Choi-5ax |
| 7eq | 34.56, CH2 | 2.48, m, 2.14, m | Choi-6 | Choi-6, Ac-2 |
| 7a | 56.45, | 4.06, m | Choi-2 | Choi-4ax,6 |
| 56.55, CH | 4.34, m | |||
| Ac 1 | 169.83, C | - | Choi-6 | - |
| 2 | 21.21, | 1.99, s | Ac-1 | Choi-7eq |
| 21.16, CH3 | 1.94, s | |||
| Ddha | - | - | - | - |
| 1 | 74.52–75.76, CH | 5.09–5.37, m | Ddha-5 | Ddha-1-OH,2,2-NH,3a,3b,7-NH2 |
| 2 | 45.28–46.83, CH | 3.76–3.97, m | Ddha-3 | Ddha-1,4,3b |
| 3a | 21.69–22.70, CH2 | 2.15–2.54, m | Ddha-2,4,5 | Ddha-1,4 |
| 3b | 1.88–2.02, m | Ddha-1,2,4,5 | Ddha-1,2,4 | |
| 4 | 107.38–108.86, CH | 5.21, 5.32, 5.16, 5.23, 5.13, 5.17, 5.21, 5.21 m | Ddha-2 | Ddha-2,3a,3b,5 |
| 5 | 121.45–122.19, CH | 6.36, d (6.7)–6.50, m | Ddha-1,3,4,7 | Ddha-4,7-NH2 |
| 1-OH | - | 6.36–6.88, brs | - | Ddha-7-NH2 |
| 2-NH | - | 7.31, m–8.39, d (7.1) | Choi-1 | Ddha-1 |
| 7 | 154.70–155.68, C | - | - | - |
| 7-NH2 | - | 7.90, brs | - | Ddha-1,1-OH,5 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan-Amer, R.; Carmeli, S. Inhibitors of Serine Proteases from a Microcystis sp. Bloom Material Collected from Timurim Reservoir, Israel. Mar. Drugs 2017, 15, 371. https://doi.org/10.3390/md15120371
Hasan-Amer R, Carmeli S. Inhibitors of Serine Proteases from a Microcystis sp. Bloom Material Collected from Timurim Reservoir, Israel. Marine Drugs. 2017; 15(12):371. https://doi.org/10.3390/md15120371
Chicago/Turabian StyleHasan-Amer, Rawan, and Shmuel Carmeli. 2017. "Inhibitors of Serine Proteases from a Microcystis sp. Bloom Material Collected from Timurim Reservoir, Israel" Marine Drugs 15, no. 12: 371. https://doi.org/10.3390/md15120371