Valorization of Lipids from Gracilaria sp. through Lipidomics and Decoding of Antiproliferative and Anti-Inflammatory Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Polar Lipidome
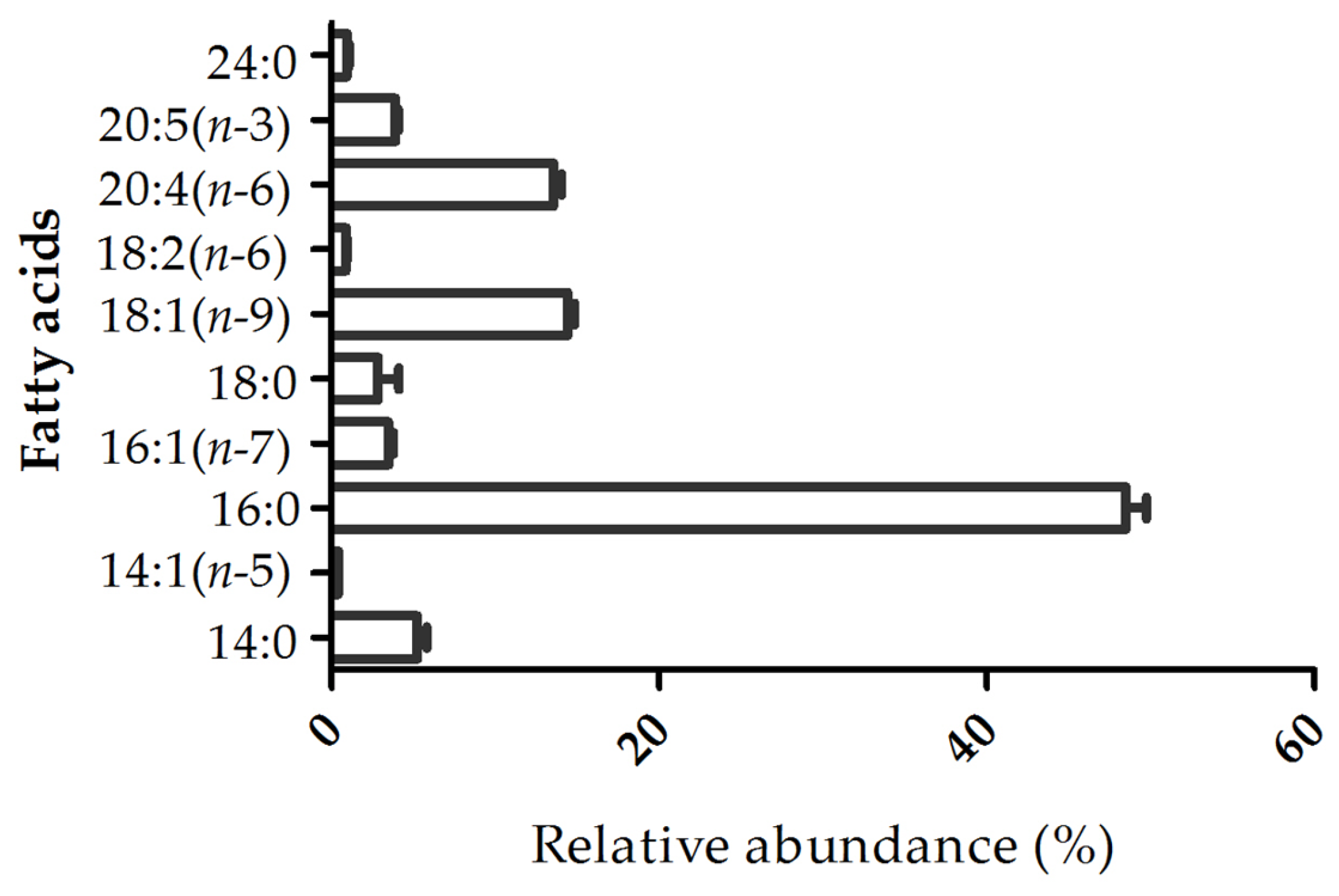
2.2. Fatty Acid Profile
2.3. Bioactivity of Lipid Extract of Gracilaria sp.
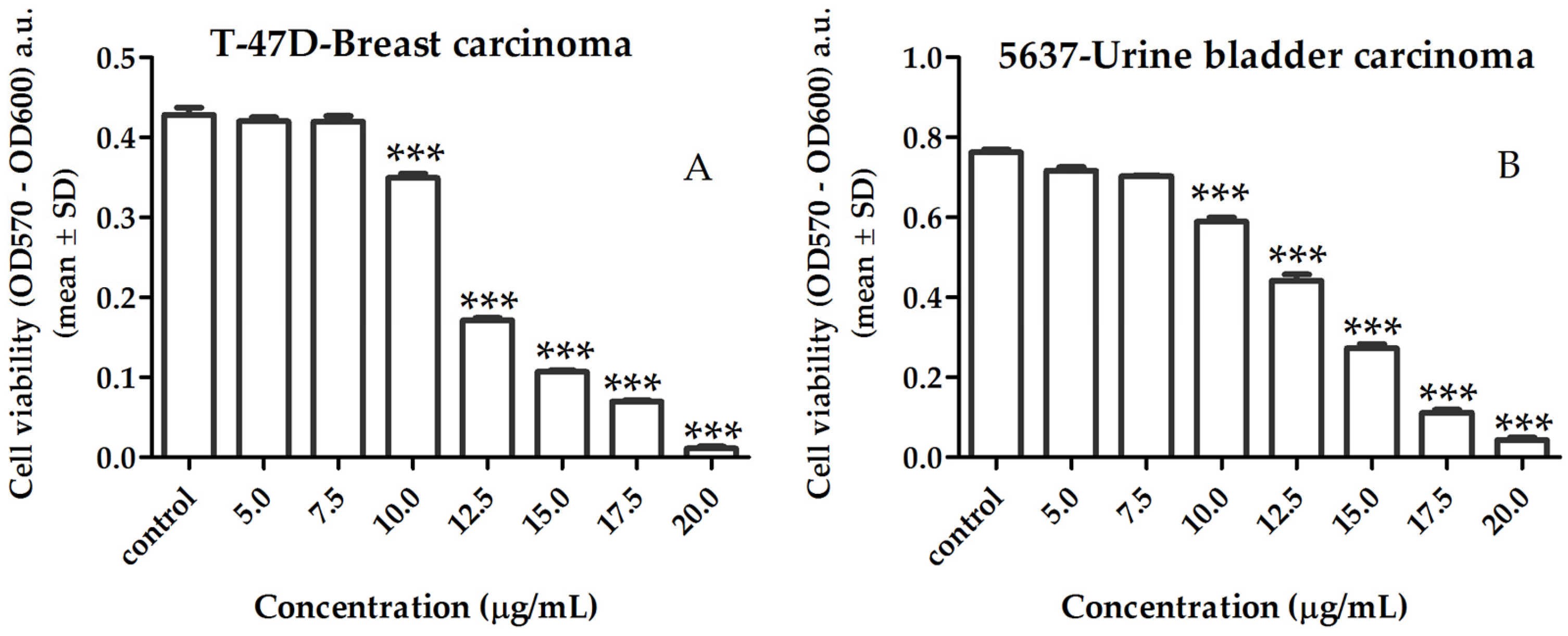
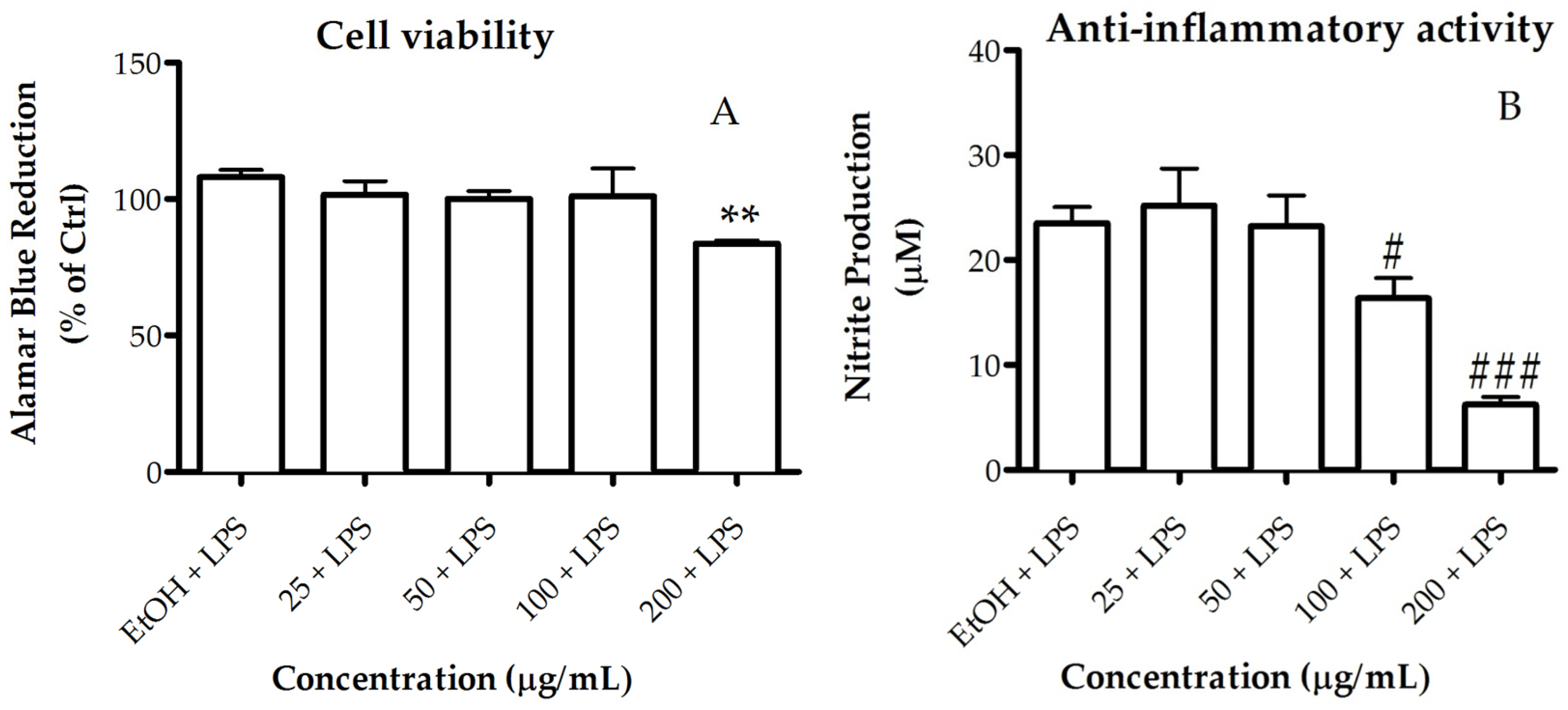
2.3.1. Activity of Lipid Extract on Human Cancer Cell Viability
2.3.2. Activity of the Lipid Extract on Nitric Oxide Production
3. Experimental Section
3.1. Biomass
3.2. Reagents
3.3. Lipid Extraction Procedure
3.4. Quantification of Glycolipids and Phospholipids
3.5. Fractionation of Lipid Extract
3.6. Hydrophilic Interaction Liquid Chromatography–Electrospray Ionization–Mass Spectrometry (HILIC–ESI–MS)
3.7. Electrospray–Mass Spectrometry (ESI–MS) Conditions
3.8. Fatty Acid Analysis by Gas Chromatography-Mass Spectrometry (GC–MS)
3.9. Cell Viability Assay on T-47D and 5637 Tumor Cell Lines
3.10. Anti-Inflammatory Activity on Nitrite Production in RAW 264.7 Cells
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Abreu, M.H.; Pereira, R.; Sassi, J.-F. Chapter 12: Marine algae and the global food industry. In Marine Algae: Biodiversity, Taxonomy, Environmental Assessment, and Biotechnology; Pereira, L., Magalhaes, J., Eds.; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Abreu, M.H.; Pereira, R.; Yarish, C.; Buschmann, A.H.; Sousa-Pinto, I. IMTA with Gracilaria vermiculophylla: Productivity and nutrient removal performance of the seaweed in a land-based pilot scale system. Aquaculture 2011, 312, 77–87. [Google Scholar] [CrossRef]
- Leal, M.C.; Rocha, R.J.M.; Rosa, R.; Calado, R. Aquaculture of marine non-food organisms: What, why and how? Rev. Aquac. 2016, 1–24. [Google Scholar] [CrossRef]
- Francavilla, M.; Franchi, M.; Monteleone, M.; Caroppo, C. The red seaweed Gracilaria gracilis as a multi products source. Mar. Drugs 2013, 11, 3754–3776. [Google Scholar] [CrossRef] [PubMed]
- Guschina, I.A.; Harwood, J.L. Lipids and lipid metabolism in eukaryotic algae. Prog. Lipid Res. 2006, 45, 160–186. [Google Scholar] [CrossRef] [PubMed]
- Van Ginneken, V.J.T.; Helsper, J.P.F.G.; de Visser, W.; van Keulen, H.; Brandenburg, W.A. Polyunsaturated fatty acids in various macroalgal species from North Atlantic and tropical seas. Lipids Health Dis. 2011, 10, 104. [Google Scholar] [CrossRef] [PubMed]
- Stengel, D.B.; Connan, S.; Popper, Z.A. Algal chemodiversity and bioactivity: Sources of natural variability and implications for commercial application. Biotechnol. Adv. 2011, 29, 483–501. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, S.; Hashim, S.N.; Rahman, H.A. Seaweeds: A sustainable functional food for complementary and alternative therapy. Trends Food Sci. Technol. 2012, 23, 83–96. [Google Scholar] [CrossRef]
- Plouguerné, E.; De Souza, L.M.; Sassaki, G.L.; Cavalcanti, J.F.; Romanos, M.T.V.; da Gama, B.A.P.; Pereira, R.C.; Barreto-Bergter, E.; Villela Romanos, M.T.; da Gama, B.A.P.; et al. Antiviral sulfoquinovosyldiacylglycerols (SQDGs) from the Brazilian brown seaweed Sargassum vulgare. Mar. Drugs 2013, 11, 4628–4640. [Google Scholar]
- Mattos, B.B.; Romanos, M.T.V.; de Souza, L.M.; Sassaki, G.; Barreto-Bergter, E. Glycolipids from macroalgae: Potential biomolecules for marine biotechnology? Rev. Bras. Farmacogn. 2011, 21, 244–247. [Google Scholar] [CrossRef]
- Plouguerné, E.; da Gama, B.A.P.; Pereira, R.C.; Barreto-Bergter, E. Glycolipids from seaweeds and their potential biotechnological applications. Front. Cell. Infect. Microbiol. 2014, 4, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Küllenberg, D.; Taylor, L.A.; Schneider, M.; Massing, U. Health effects of dietary phospholipids. Lipids Health Dis. 2012, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, E.; Silva, J.; Mendonça, S.; Abreu, M.; Domingues, M. Lipidomic approaches towards deciphering glycolipids from microalgae as a reservoir of bioactive lipids. Mar. Drugs 2016, 14, 101. [Google Scholar] [CrossRef] [PubMed]
- Maciel, E.; Leal, M.C.; Lillebø, A.I.; Domingues, P.; Domingues, M.R.; Calado, R. Bioprospecting of marine macrophytes using MS-based lipidomics as a new approach. Mar. Drugs 2016, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Robertson, R.C.; Guihéneuf, F.; Bahar, B.; Schmid, M.; Stengel, D.B.; Fitzgerald, G.F.; Ross, R.P.; Stanton, C. The Anti-Inflammatory effect of algae-derived lipid extracts on lipopolysaccharide (LPS)-stimulated human THP-1 macrophages. Mar. Drugs 2015, 13, 5402–5424. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.; Pan, B.S. Identification of sulfoglycolipid bioactivities and characteristic fatty acids of marine macroalgae. JAFC 2012, 60, 8404–8410. [Google Scholar] [CrossRef] [PubMed]
- Ohta, K.; Mizushina, Y.; Hirata, N.; Takemura, M.; Sugawara, F.; Matsukage, A.; Yoshida, S.; Sakaguchi, K. Sulfoquinovosyldiacylglycerol, KM043, a new potent inhibitor of eukaryotic DNA polymerases and HIV-reverse transcriptase type 1 from a marine red alga, Gigartina tenella. Chem. Pharm. Bull. 1998, 46, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Bergé, J.P.; Debiton, E.; Dumay, J.; Durand, P.; Barthomeuf, C. In vitro anti-inflammatory and anti-proliferative activity of sulfolipids from the red alga Porphyridium cruentum. J. Agric. Food Chem. 2002, 50, 6227–6232. [Google Scholar] [CrossRef] [PubMed]
- Lopes, G.; Daletos, G.; Proksch, P.; Andrade, P.B.; Valentão, P. Anti-inflammatory potential of monogalactosyl diacylglycerols and a monoacylglycerol from the edible brown seaweed Fucus spiralis Linnaeus. Mar. Drugs 2014, 12, 1406–1418. [Google Scholar] [CrossRef] [PubMed]
- Banskota, A.H.; Stefanova, R.; Sperker, S.; Lall, S.P.; Craigie, J.S.; Hafting, J.T.; Critchley, A.T. Polar lipids from the marine macroalgae Palmaria palmata inhibit lipopolysaccharide-induced nitric oxide production in RAW264.7 macrophage cells. Phytochemistry 2014, 101, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Banskota, A.H.; Stefanova, R.; Sperker, S.; Lall, S.; Craigie, J.S.; Hafting, J.T. Lipids isolated from the cultivated red alga Chondrus crispus inhibit nitric oxide production. J. Appl. Phycol. 2014, 26, 1565–1571. [Google Scholar] [CrossRef]
- Maeda, N.; Kokai, Y.; Ohtani, S.; Sahara, H.; Hada, T.; Ishimaru, C.; Kuriyama, I.; Yonezawa, Y.; Iijima, H.; Yoshida, H.; et al. Anti-tumor effects of the glycolipids fraction from spinach which inhibited DNA polymerase activity. Nutr. Cancer 2007, 57, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.S.; Rodríguez, A.D.; Berlinck, R.G.S.; Fusetani, N. Marine pharmacology in 2007–2008: Marine compounds with antibacterial, anticoagulant, antifungal, anti-inflammatory, antimalarial, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous system, and other miscellaneous mec. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011, 153, 191–222. [Google Scholar] [PubMed]
- Pereira, H.; Barreira, L.L.; Figueiredo, F.; Custódio, L.L.; Vizetto-Duarte, C.; Polo, C.; Rešek, E.; Engelen, A.; Varela, J.J. Polyunsaturated fatty acids of marine macroalgae: Potential for nutritional and pharmaceutical applications. Mar. Drugs 2012, 10, 1920–1935. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, E.; Melo, T.; Moreira, A.S.P.; Alves, E.; Domingues, P.; Calado, R.; Abreu, M.H.M.H.; Domingues, M.R. Decoding bioactive polar lipid profile of the macroalgae Codium tomentosum from a sustainable IMTA system using a lipidomic approach. Algal Res. 2015, 12, 388–397. [Google Scholar] [CrossRef]
- Melo, T.; Alves, E.; Azevedo, V.V.; Martins, A.S.; Neves, B.; Domingues, P.; Calado, R.; Abreu, M.H.; Domingues, M.R. Lipidomics as a new approach for the bioprospecting of marine macroalgae-Unraveling the polar lipid and fatty acid composition of Chondrus crispus. Algal Res. 2015, 8, 181–191. [Google Scholar] [CrossRef]
- Dembitsky, V.M.; Řezanková, H.; Řezanka, T.; Hanuš, L.O. Variability of the fatty acids of the marine green algae belonging to the genus Codium. Biochem. Syst. Ecol. 2003, 31, 1125–1145. [Google Scholar] [CrossRef]
- Khotimchenko, S.Y.V.; Vaskovsky, Y.E.; Titlyanova, T.V.; Vaskovsky, V.E.; Titlyanova, T.V. Fatty acids of marine algae from the Pacific Coast of North California. Bot. Mar. 2002, 45, 17–22. [Google Scholar] [CrossRef]
- Ragonese, C.; Tedone, L.; Beccaria, M.; Torre, G.; Cichello, F.; Cacciola, F.; Dugo, P.; Mondello, L. Characterisation of lipid fraction of marine macroalgae by means of chromatography techniques coupled to mass spectrometry. Food Chem. 2014, 145, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Khotimchenko, S.V. Distribution of glyceroglycolipids in marine algae and grasses. Chem. Nat. Compd. 2002, 38, 186–191. [Google Scholar] [CrossRef]
- Khotimchenko, S.V. Lipids from marine alga Gracilaria verrucosa. Chem. Nat. Compd. 2005, 41, 230–232. [Google Scholar] [CrossRef]
- Sanina, N.M.; Goncharova, S.N.; Kostetsky, E.Y. Fatty acid composition of individual polar lipid classes from marine macrophytes. Phytochemistry 2004, 65, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Kendel, M.; Couzinet-Barnathan, G.; Mossion, A.; Viau, M.; Fleurence, J.; Barnathan, G.; Wielgosz-Collin, G. Seasonal composition of lipids, fatty acids, and sterols in the edible red alga Grateloupia turuturu. J. Appl. Phycol. 2013, 25, 425–432. [Google Scholar] [CrossRef]
- He, H.; Rodgers, R.P.; Marshall, A.G.; Hsu, C.S. Algae polar lipids characterized by online liquid chromatography coupled with hybrid linear quadrupole ion trap/fourier transform ion cyclotron resonance mass spectrometry. Energy Fuels 2011, 25, 4770–4775. [Google Scholar] [CrossRef]
- Naumann, I.; Darsow, K.H.; Walter, C.; Lange, H.A.; Buchholz, R. Identification of sulfoglycolipids from the alga Porphyridium purpureum by matrix-assisted laser desorption/ionisation quadrupole ion trap time-of-flight mass spectrometry. Rappid Commun. Mass Spectrom. 2007, 21, 3185–3192. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Kamide, Y.; Hirai, M.Y.; Saito, K. Plant lipidomicss based on hydrophilic interaction chromatography coupled to ion trap time-of flight mass spectrometry. Metabolomics. 2011, 9, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Kumar, M.; Reddy, C.R.K.; Jha, B. Nitrate and phosphate regimes induced lipidomic and biochemical changes in the intertidal macroalga Ulva lactuca (Ulvophyceae, Chlorophyta). Plant Cell Physiol. 2014, 55, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Popendorf, K.J.; Fredricks, H.F.; Van Mooy, B.A.S. Molecular ion-independent quantification of polar glycerolipid classes in marine plankton using triple quadrupole MS. Lipids 2013, 48, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Leal, M.C.; Munro, M.H.G.; Blunt, J.W.; Puga, J.; Jesus, B.; Calado, R.; Rosa, R.; Madeira, C. Biogeography and biodiscovery hotspots of macroalgal marine natural products. Nat. Prod. Rep. 2013, 30, 1380–1390. [Google Scholar] [CrossRef] [PubMed]
- Pettitt, T.R.; Harwood, J.L. Alterations in lipid metabolism caused by illumination of the marine red algae Chondrus crispus and Polysiphonia lanosa. Phytochemistry 1989, 28, 3295–3300. [Google Scholar] [CrossRef]
- Trevor, R.; Pettitt, A.; Jones, L.; Harwood, J.L. Lipids of the marine red algae, Chondrus crispus and Polysiphonia lanosa. Phytochemistry 1989, 28, 399–405. [Google Scholar] [CrossRef]
- Harwood, J.L. Lipid metabolism in the red marine algae Chondrus Crispus and Polysinphonza Lanosa as modified by temperature. Phytochemistry 1989, 28, 1–6. [Google Scholar]
- De Souza, L.M.; Sassaki, G.L.; Romanos, M.T.V.; Barreto-Bergter, E. Structural characterization and anti-HSV-1 and HSV-2 activity of glycolipids from the marine algae Osmundaria obtusiloba isolated from Southeastern Brazilian coast. Mar. Drugs 2012, 10, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Khotimchenko, S.V.; Vaskovsky, V.E. An inositol-containing sphingolipid from the red alga Gracilaria verrucosa. Russ. J. Bioorg. Chem. 2004, 30, 168–171. [Google Scholar] [CrossRef]
- Sato, N. Betaine Lipids. Bot. Mag. 1992, 1, 185–197. [Google Scholar] [CrossRef]
- Dembitsky, V.M. Betaine ether-linked glycerolipids. Prog. Lipid Res. 1996, 35, 1–51. [Google Scholar] [CrossRef]
- Khotimchenko, S.V. Variations in lipid composition among different developmental stages of Gracilaria verrucosa (Rhodophyta). Bot. Mar. 2006, 49, 34–38. [Google Scholar] [CrossRef]
- Diet, Nutrition and the Prevention of Chronic Diseases; WHO Technical Report Series 916; World Health Organization: Geneva, Switzerland, 2003; Available online: http://www.who.int/dietphysicalactivity/publications/trs916/en/ (accessed on 10 January 2017).
- Simopoulos, A. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. 2008, 233, 674–688. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A. The importance of the ratio of omega-6/omega-3 essential fatty acids. Biomed. Pharmacother. 2002, 56, 365–379. [Google Scholar] [CrossRef]
- Husted, K.S.; Bouzinova, E.V. The importance of n-6/n-3 fatty acids ratio in the major depressive disorder. Medicina 2016, 52, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.; Hraiki, A.; Bebawy, M.; Pazderka, C.; Rawling, T. Anti-tumor activities of lipids and lipid analogues and their development as potential anticancer drugs. Pharmacol. Ther. 2015, 150, 109–128. [Google Scholar] [CrossRef] [PubMed]
- Hossain, Z.; Kurihara, H.; Hosokawa, M.; Takahashi, K. Growth inhibition and induction of differentiation and apoptosis mediated by sodium butyrate in Caco-2 cells with algal glycolipids. In Vitro Cell. Dev. Biol. Anim. 2005, 41, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, C.; Yu, G.; Guan, H. Total synthesis and structure-activity relationship of glycoglycerolipids from marine organisms. Mar. Drugs 2014, 12, 3634–3659. [Google Scholar] [CrossRef] [PubMed]
- Burri, L.; Hoem, N.; Banni, S.; Berge, K. Marine omega-3 phospholipids: Metabolism and biological activities. Int. J. Mol. Sci. 2012, 13, 15401–15419. [Google Scholar] [CrossRef] [PubMed]
- D’Arrigo, P.; Servi, S. Synthesis of lysophospholipids. Molecules 2010, 15, 1354–1377. [Google Scholar] [CrossRef] [PubMed]
- Banskota, A.H.; Gallant, P.; Stefanova, R.; Melanson, R.; O’Leary, S.J.B. Monogalactosyldiacylglycerols, potent nitric oxide inhibitors from the marine microalga Tetraselmis chui. Nat. Prod. Res. 2012, 27, 37–41. [Google Scholar]
- Koch, A.K.; Kappeli, O.; Fiechter, A.; Reiser, J. Hydrocarbon assimilation and biosurfactant production in Pseudomonas aeruginosa mutants. J. Bacteriol. 1991, 173, 4212–4219. [Google Scholar] [CrossRef] [PubMed]
- Dória, M.L.; Cotrim, Z.; Macedo, B.; Simões, C.; Domingues, P.; Helguero, L.; Domingues, M.R. Lipidomic approach to identify patterns in phospholipid profiles and define class differences in mammary epithelial and breast cancer cells. Breast Cancer Res. Treat. 2012, 133, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, E.M.; Lewis, D.H. Spectrophotometric determination of phosphate esters in the presence and absence of orthophosphate. Anal. Biochem. 1970, 36, 159–167. [Google Scholar] [CrossRef]
- Pacetti, D.; Boselli, E.; Lucci, P.; Frega, N.G. Simultaneous analysis of glycolipids and phospholids molecular species in avocado (Persea americana Mill) fruit. J. Chromatogr. A 2007, 1150, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Melo, T.; Silva, E.M.P.; Simões, C.; Domingues, P.; Domingues, M.R.M. Photooxidation of glycated and non-glycated phosphatidylethanolamines monitored by mass spectrometry. J. Mass Spectrom. 2013, 48, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Dória, M.L.; Cotrim, C.Z.; Simões, C.; Macedo, B.; Domingues, P.; Domingues, M.R.; Helguero, L.A. Lipidomic analysis of phospholipids from human mammary epithelial and breast cancer cell lines. J. Cell. Physiol. 2013, 228, 457–468. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef] [PubMed]
- Green, L.C.; Wagner, D.A.; Glogowski, J.; Skipper, P.L.; Wishnok, J.S.; Tannenbaum, S.R. Analysis of nitrate, nitrite, and 15N nitrate in biological fluids. Anal. Biochem. 1982, 126, 131–138. [Google Scholar] [CrossRef]
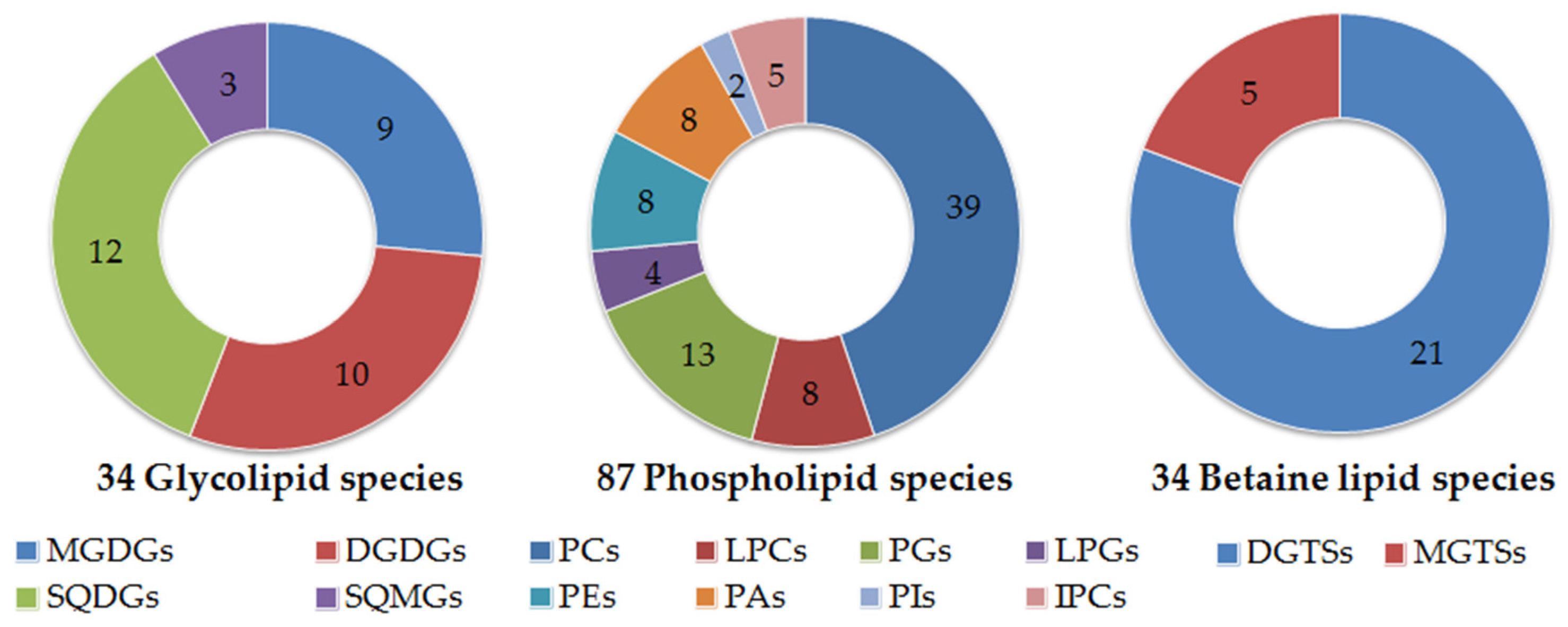
| Composition | Mean | SD |
|---|---|---|
| Lipids (mg/kg biomass) | 3000 | 600 |
| Glycolipids (mg/kg biomass) | 1980 | 148 |
| Phospholipids (mg/kg biomass) | 165 | 52.7 |
| Betaines and others 1 | 855 | - |
| [M + NH4]+ | Lipid Species | Fatty Acyl Chains |
| m/z | (C:N) | |
| Monogalactosyl diacylglyceride (MGDG) | ||
| 746.3 | MGDG (32:1) | 16:1/16:0 and 14:0/18:1 |
| 748.3 | MGDG (32:0) | 16:0/16:0 and 14:0/18:0 |
| 774.3 | MGDG (34:1) | 18:1/16:0 |
| 776.3 | MGDG (34:0) | 18:0/16:0 |
| 794.3 | MGDG (36:5) | 20:5/16:0 |
| 796.3 | MGDG (36:4) | 20:4/16:0 and 18:2/18:2 |
| Digalactosyl diacylglyceride (DGDG) | ||
| 908.3 | DGDG (32:1) | 16:1/16:0 and 14:0/18:1 |
| 910.3 | DGDG (32:0) | 16:0/16:0 and 14:0/18:0 |
| 934.3 | DGDG (34:2) | 18:2/16:0 and 18:1/16:1 |
| 936.3 | DGDG (34:1) | 18:1/16:0 |
| 956.3 | DGDG (36:5) | 20:5/16:0 |
| 958.3 | DGDG (36:4) | 20:4/16:0 and 18:2/18:2 |
| [M − H]− | Lipid Species | Fatty Acyl Chains |
| Sulfoquinovosyl diacylglyceride (SQDG) | ||
| 763.6 | SQDG (30:1) | 14:0/16:1 |
| 765.6 | SQDG (30:0) | 14:0/16:0 |
| 791.6 | SQDG (32:1) | 16:1/16:0 and 14:0/18:2 |
| 793.6 | SQDG (32:0) | 16:0/16:0 and 14:0/18:0 |
| 813.6 | SQDG (34:4) | 18:4/16:0 |
| 817.6 | SQDG (34:2) | 18:2/16:0 |
| 819.6 | SQDG (34:1) | 18:1/16:0 |
| 839.6 | SQDG (36:5) | 20:5/16:0 |
| 841.6 | SQDG (36:4) | 20:4/16:0 |
| 857.6 | SQDG (36:4-OH) | 20:4-OH/16:0 |
| Sulfoquinovosyl monoacylglyceride (SQMG) | ||
| 527.4 | SQMG (14:0) | |
| 553.4 | SQMG (16:1) | |
| 555.4 | SQMG (16:0) | |
| [M + H]+ | Lipid Species | Fatty Acyls Chain |
| m/z | (C:N) | |
| Phosphatidylcholine (PC) | ||
| 732.6 | PC (32:1) | 16:0/16:1 and 14:0/18:1 |
| 734.6 | PC (32:0) | 16:0/16:0 and 14:0/18:0 |
| 754.6 | PC (34:4) | 14:0/20:4 and 16:2/18:2 |
| 756.6 | PC (34:3) | 16:0/18:3 and 14:0/20:3 |
| 758.6 | PC (34:2) | 16:0/18:2 and 16:2/18:1 |
| 760.6 | PC (34:1) | 16:0/18:1 |
| 762.6 | PC (34:0) | 16:0/18:0 |
| 780.6 | PC (36:5) | 16:0/20:5 and 18:2/18:3 |
| 782.6 | PC (36:4) | 16:0/20:4 and 18:2/18:2 |
| 784.6 | PC (36:3) | 16:0/20:3 and 18:1/18:2 |
| 786.6 | PC (36:2) | 18:0/18:2 and 18:1/18:1 |
| 788.6 | PC (36:1) | 18:0/18:1 |
| 798.5 | PC (37:3) | 16:0/21:3 and 18:1/19:2 |
| 804.5 | PC (38:7) | 18:3/20:4 and 18:2/20:5 |
| 806.5 | PC (38:6) | 18:2/20:4 and 18:1/20:5 |
| 808.5 | PC (38:5) | 18:1/20:4 and 18:2/20:3 |
| 810.5 | PC (38:4) | 18:1/20:3 and 16:0/22:4 |
| 812.5 | PC (38:3) | 18:0/20:3 and 18:1/20:2 |
| 814.5 | PC (38:2) | 16:0/22:2 and 18:1/20:1 |
| 818.5 | PC (38:0) | 18:0/20:0 and 16:0/22:0 |
| 840.4 | PC (40:3) | 18:1/22:2 |
| 844.4 | PC (40:1) | 18:1/22:0 |
| Lyso-phosphatidylcholine (LPC) | ||
| 494.4 | LPC (16:1) | |
| 496.4 | LPC (16:0) | |
| 518.4 | LPC (18:3) | |
| 520.4 | LPC (18:2) | |
| 522.4 | LPC (18:1) | |
| 524.4 | LPC (18:0) | |
| 542.4 | LPC (20:5) | |
| 544.4 | LPC (20:4) | |
| Phosphatidyletanolamine (PE) | ||
| 716.4 | PE (34:2) | 16:1/18:1 and 16:0/18:2 |
| 718.3 | PE (34:1) | 16:1/18:0 and 16:0/18:1 |
| 740.4 | PE (34:0) | 16:0/18:0 |
| 742.4 | PE (36:3) | 18:1/18:2 |
| 744.4 | PE (36:2) | 18:1/18:1 |
| 746.3 | PE (36:1) | 18:0/18:1 |
| [M − H]− | Lipid Species | Fatty Acyl Chains |
| Phosphatidylglycerol (PG) | ||
| 717.4 | PG (32:2) | 16:1/16:1 and 16:0/16:2 |
| 719.4 | PG (32:1) | 16:0/16:1 |
| 721.4 | PG (32:0) | 16:0/16:0 |
| 741.4 | PG (34:4) | 16:0/18:4 |
| 743.5 | PG (34:3) | 16:0/18:3 |
| 745.5 | PG (34:2) | 16:1/18:1 |
| 747.5 | PG (34:1) | 16:0/18:1 and 16:1/18:0 |
| 767.5 | PG (36:5) | 16:0/20:5 |
| 769.4 | PG (36:4) | 16:0/20:4 and 18:2/18:2 |
| 773.5 | PG (36:2) | 18:1/18:1 |
| Lyso-phosphatidylglycerol (LPG) | ||
| 481.3 | LPG (16:1) | |
| 483.3 | LPG (16:0) | |
| 509.3 | LPG (18:1) | |
| 531.3 | LPG (20:4) | |
| Phosphatidylinositol (PI) | ||
| 833.5 | PI (34:2) | 16:1/18:1 |
| 835.5 | PI (34:1) | 16:0/18:1 |
| Phosphatidic acid (PA) | ||
| 693.4 | PA (36:5) | 16:0/20:5 |
| 695.4 | PA (36:4) | 16:0/20:4 |
| 717.4 | PA (38:7) | 18:3/20:4 |
| 719.4 | PA (38:6) | 18:2/20:4 |
| 721.4 | PA (38:5) | 18:1/20:4 |
| 741.3 | PA (40:9) | 20:4/20:5 |
| 743.3 | PA (40:8) | 20:4/20:4 |
| 745.3 | PA (40:7) | 20:3/20:4 |
| Inositolphosphoceramide (IPC) | ||
| 810.5 | IPC (t35:0) | t18:0/17:0 |
| 908.6 | IPC (d42:0) | d18:0/24:0 |
| 920.6 | IPC (t42:2) | t18:1/24:1 |
| 922.6 | IPC (t42:1) | t18:0/24:1 |
| 924.6 | IPC (t42:0) | t18:0/24:0 |
| [M + H]+ | Lipid Species | Fatty Acyls Chain |
| m/z | (C:N) | |
| Diacylglyceryl trimethyl homoserine (DGTS) | ||
| 656.7 | DGTS (28:0) | 14:0/14:0 |
| 682.7 | DGTS (30:1) | 14:0/16:1 |
| 684.8 | DGTS (30:0) | 14:0/16:0 |
| 708.7 | DGTS (32:2) | 16:1/16:1 and 14:0/18:2 |
| 710.7 | DGTS (32:1) | 16:0/16:1 and 14:0/18:1 |
| 712.7 | DGTS (32:0) | 16:0/16:0 and 14:0/18:0 |
| 732.7 | DGTS (34:4) | 16:2/18:2 and 14:0/20:4 |
| 734.7 | DGTS (34:3) | 16:1/18:2 |
| 736.7 | DGTS (34:2) | 16:0/18:2 and 16:1/18:1 |
| 738.7 | DGTS (34:1) | 16:0/18:1 and 16:1/18:0 |
| 740.7 | DGTS (34:0) | 16:0/18:0 and 14:0/20:0 |
| 760.6 | DGTS (36:4) | 16:0/20:4 |
| 764.8 | DGTS (36:2) | 18:1/18:1 |
| 766.8 | DGTS (36:1) | 18:0/18:1 |
| Monoacylglyceryl trimethyl homoserine (MGTS) | ||
| 446.5 | MGTS (14:0) | |
| 472.5 | MGTS (16:1) | |
| 474.5 | MGTS (16:0) | |
| 498.6 | MGTS (18:2) | |
| 500.6 | MGTS (18:1) | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Da Costa, E.; Melo, T.; Moreira, A.S.P.; Bernardo, C.; Helguero, L.; Ferreira, I.; Cruz, M.T.; Rego, A.M.; Domingues, P.; Calado, R.; et al. Valorization of Lipids from Gracilaria sp. through Lipidomics and Decoding of Antiproliferative and Anti-Inflammatory Activity. Mar. Drugs 2017, 15, 62. https://doi.org/10.3390/md15030062
Da Costa E, Melo T, Moreira ASP, Bernardo C, Helguero L, Ferreira I, Cruz MT, Rego AM, Domingues P, Calado R, et al. Valorization of Lipids from Gracilaria sp. through Lipidomics and Decoding of Antiproliferative and Anti-Inflammatory Activity. Marine Drugs. 2017; 15(3):62. https://doi.org/10.3390/md15030062
Chicago/Turabian StyleDa Costa, Elisabete, Tânia Melo, Ana S. P. Moreira, Carina Bernardo, Luisa Helguero, Isabel Ferreira, Maria Teresa Cruz, Andreia M. Rego, Pedro Domingues, Ricardo Calado, and et al. 2017. "Valorization of Lipids from Gracilaria sp. through Lipidomics and Decoding of Antiproliferative and Anti-Inflammatory Activity" Marine Drugs 15, no. 3: 62. https://doi.org/10.3390/md15030062