Isolation and Selection of Microalgal Strains from Natural Water Sources in Viet Nam with Potential for Edible Oil Production
Abstract
:1. Introduction
2. Results
2.1. Isolation of Lipid-Producing Microalgal Strains
2.2. Molecular Characterization of Microalgal Strains
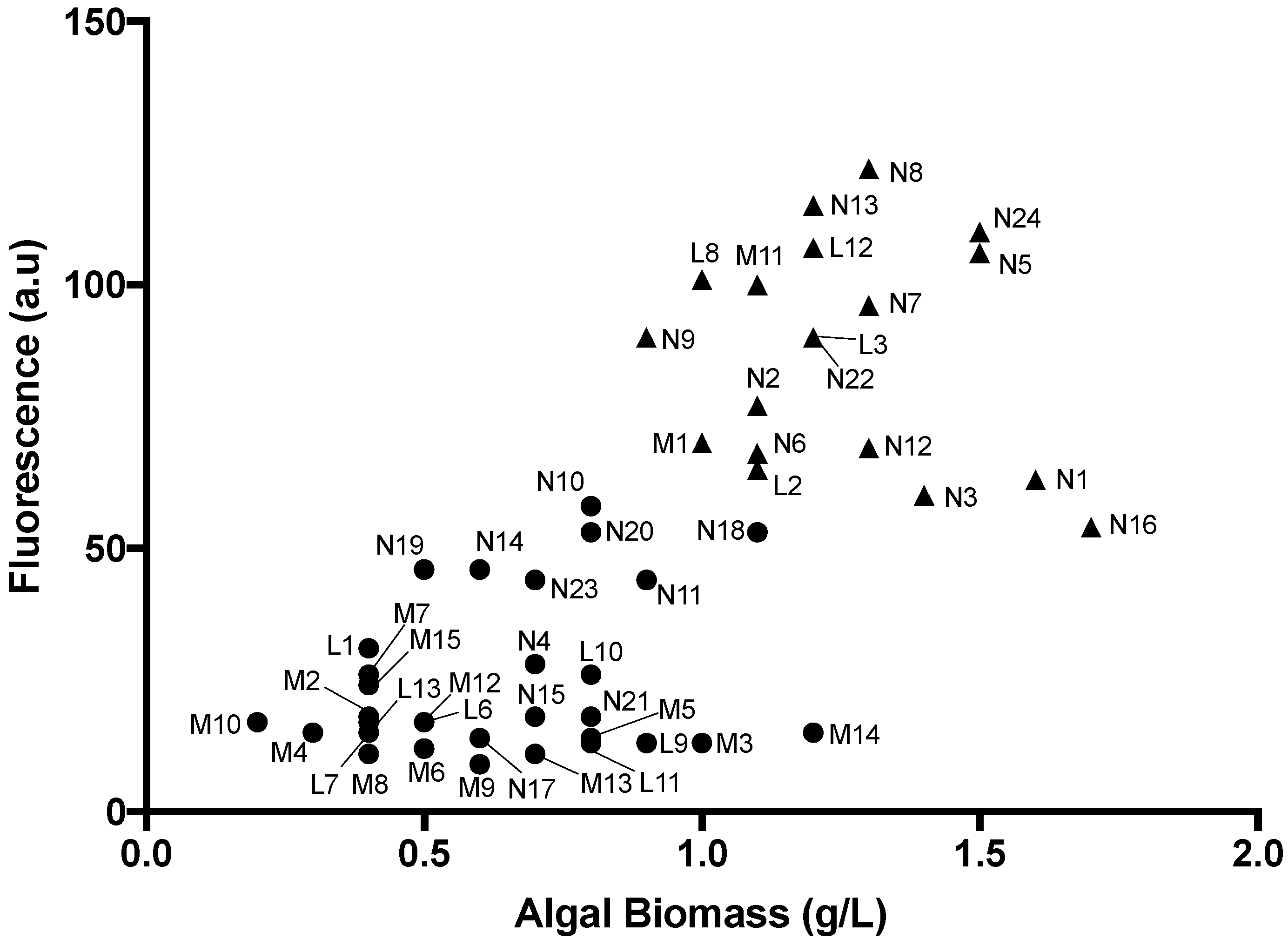
2.3. Identification of High Lipid-and High Biomass-Producing Microalgae
2.4. Fatty Acid Composition of Microalgal Lipids
3. Discussion
4. Materials and Methods
4.1. Water Sampling
4.2. Isolation of Lipid-Producing Microalgae
4.3. Detection of Lipid-Producing Microalgae
4.4. Sequencing of 18S RNA Genes
4.5. Screening of Microalgae for High Biomass Production
4.6. Lipid Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- General Statistics Office of Viet Nam; Statistical Documentation and Service Centre: Ha Noi, Vietnam, 2015.
- Vietnam Trade Promotion Agency. Production and Consumption of Vegetable Oils in Viet Nam to 2025. Available online: www.vietrade.gov.vn (accessed on 25 June 2014).
- Chisti, Y. Biodiesel from microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Sydney, E.B.; Sturm, W.; de Carvalho, J.C.; Thomaz-Soccol, V.; Larroche, C.; Pandey, A.; Soccol, C.R. Potential carbon dioxidefixation by industrially important microalgae. Bioresour. Technol. 2010, 101, 5892–5896. [Google Scholar] [CrossRef] [PubMed]
- Um, B.H.; Kim, Y.S. Review: A chance for Korea to advance algal-biodiesel technology. J. Ind. Eng. Chem. 2009, 15, 1–7. [Google Scholar] [CrossRef]
- Illman, A.M.; Scragg, A.H.; Shales, S.W. Increase in Chlorella strains calorific values when grown in low nitrogen medium. Enzyme Microb. Technol. 2000, 27, 631–635. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Wang, G.C.; Zhou, B.C. Effect of iron on growth and lipid accumulation in Chlorella vulgaris. Bioresour. Technol. 2007, 99, 4717–4722. [Google Scholar] [CrossRef] [PubMed]
- Spolaore, P.; Joannis-Cassan, C.; Duran, E.; Isambert, A. Commercial applications of microalgae. J. Biosci. Bioeng. 2006, 101, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Mata, T.M.; Martins, A.A.; Caetano, N.S. Microalgae for biodiesel production and other applications: A review. Renew. Sustain. Energy Rev. 2010, 14, 217–232. [Google Scholar] [CrossRef]
- Hồng, D.D. Establishing an Assemblage of Marine Photoautotrophic and Heterotrophic Microalgae in Viet Nam and Growing Some Heterotrophic Strains for Biomass for Aqua-Culture; Final Report, Biotechnology Program for Agriculture and Aqua-Culture; Ministry of Agriculture and Rural Development: Ha Noi, Vietnam, 2010. (In Vietnamese)
- Lan, N.T.M. Selection and Improvement of Lipid Accumulation of Microalgae, Background for Biodiesel Production. Ph.D. Thesis, Natural University of Ho Chi Minh City, Ho Chi Minh City, VietNam, 2013. (In Vietnamese). [Google Scholar]
- Sơn, D.T. Initial Observation about Lipids and Nutrients of Some Microalgal Strains Originated in Viet Nam. J. Sci. Technol. Inf. (STNFO) 2014, 11, 29–31. (In Vietnamese) [Google Scholar]
- Kirst, G.O. Salinity tolerance of eukaryotic marine algae. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 21–53. [Google Scholar] [CrossRef]
- Estrada, M.; Henriksen, P.; Gasol, J.M.; Casamayor, E.O. Diversity of planktonic photoautotrophic microorganisms along a salinity gradient as depicted by microscopy, flow cytometry, pigment analysis and DNA-based methods. FEMS Microbiol. Ecol. 2004, 49, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Qin, B.; Paerl, H.W.; Zhang, Y.; Wu, P.; Ma, J.; Chen, Y. Effects of nutrients, temperature and their interactions on Spring phytoplankton community succession in Lake Taihu, China. PLoS ONE 2014, 9, e113960. [Google Scholar] [CrossRef] [PubMed]
- Tilman, D.; Kilham, S.S.; Kilham, P. Phytoplankton community ecology: The role of limiting nutrients. Ann. Rev. Ecol. Syst. 1982, 13, 349–372. [Google Scholar] [CrossRef]
- Rodolfi, L.; Zittelli, G.C.; Bassi, N.; Padovani, G.; Biondi, N.; Bonini, G.; Tredici, M.R. Microalgae for oil: Strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 2009, 102, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Ajjawi, I.; Verruto, J.; Aqui, M.; Soriaga, L.B.; Coppersmith, J.; Kwok, K.; Peach, L.; Orchard, E.; Kalk, R.; Xu, W.; et al. Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat. Biotechnol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Chiu, S.Y.; Kao, C.Y.; Tsai, M.T.; Ong, S.C.; Chen, C.H.; Lin, C.S. Lipid accumulation and CO2 utilization of Nannochloropsis oculata in response to CO2 aeration. Bioresour. Technol. 2009, 100, 833–838. [Google Scholar] [CrossRef] [PubMed]
- De Morais, M.G.; Costa, J.A.V. Carbon dioxide fixation by Chlorella kessleri, C. vulgaris, Scenedesmus obliquus and Spirulina sp. cultivated in flasks and vertical tubular photobioreactors. Biotechnol. Lett. 2007, 29, 1349–1352. [Google Scholar] [CrossRef] [PubMed]
- Gouveia, L.; Oliveira, A.C. Microalgae as a raw material for biofuels production. J. Ind. Microbiol. Biotechnol. 2009, 36, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Ugwu, C.U.; Aoyagi, H.; Uchiyama, H. Photobioreactors for mass cultivation of algae. Bioresour. Technol. 2008, 99, 4021–4028. [Google Scholar] [CrossRef] [PubMed]
- Zambiazi, R.C.; Przybylski, R.; Zambiazi, M.W.; Mendonca, C.B. Fatty acid composition of vegetable oils and fats. Bol. CEPPA Curitiba 2007, 25, 111–120. [Google Scholar]
- Dauqan, E.; Sani, H.A.; Abdullah, A.; Kasim, Z.M. Effect of different vegetable oils (red palm olein, palm lein, corn oil and coconut oil) on lipid profile in rat. Food Nut. Sci. 2011, 2, 253–258. [Google Scholar] [CrossRef]
- Widjaja, A. Study of increasing lipid production from fresh water microalgae Chlorella vulgaris. J. Taiwan Inst. Chem. E 2009, 40, 13–20. [Google Scholar] [CrossRef]
- Raghavan, G.; Haridevi, C.K.; Gopinathan, C.P. Growth and proximate composition of the Chaetoceros calcitrans f. Pumilus under different temperature, salinity and carbon dioxide levels. Aquac. Res. 2008, 39, 1053–1058. [Google Scholar] [CrossRef]
- Chader, S.; Mahmah, B.; Chetehouna, K.; Mignolet, E. Biodiesel production using Chlorella sorokiniana a green microalga. Rev. Energies Renouv. 2011, 14, 21–26. [Google Scholar]
- Becker, E.W. Culture Media. In Microalgae: Biotechnology and Microbiology; Becker, E.W., Ed.; Cambridge University Press: Cambridge, UK, 1994; pp. 9–41. [Google Scholar]
- Metting, F. Biodiversity and application of microalgae. J. Ind. Microbiol. Biotechnol. 1996, 17, 477–489. [Google Scholar] [CrossRef]
- Stanier, R.V.; Kunisawa, R.; Mandel, M.; Cohen-Bazire, G. Purification and properties of unicellular blue-green algae (order: Chrococcales). Bacteriol. Rev. 1971, 35, 171–205. [Google Scholar] [PubMed]
- Guillard, R.L.; Ryther, J.H. Studies on marine planktonic diatoms: Cyclotella nana Hustedt and Detonula confevacea (Cleve). Gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Cooksey, K.E.; Guckert, J.B.; Williams, S.A.; Callis, P.R. Fluorometric determination of the neutral lipid-content of microalgal cells using Nile Red. J. Microbiol. Meth. 1987, 6, 333–345. [Google Scholar] [CrossRef]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Naghidi, F.G.; Garg, S.; Adarme-Vega, T.C.; Thurecht, K.J.; Ghafor, W.A.; Tannock, S.; Schenk, P.M. A comparative study: The impact of different lipid extraction methods on current microalgal lipid research. Microb. Cell Factories 2014, 13, 14. [Google Scholar] [CrossRef] [PubMed]
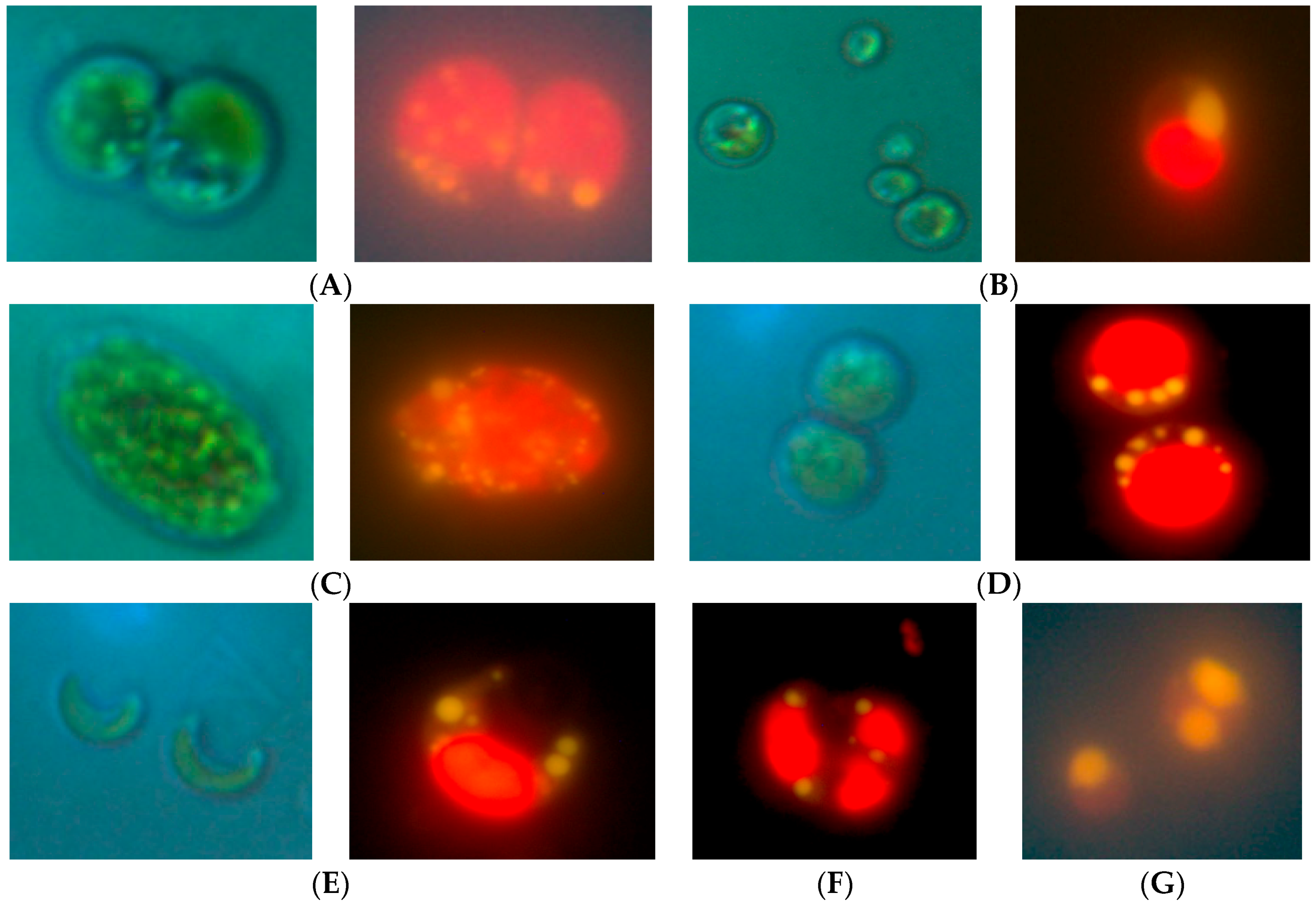

| No. | Name | Species | Nearest Relative | GenBank Number of Relative |
|---|---|---|---|---|
| 1 | N1 * | Chlamydomonas sp. | 99% | FR865562.1 |
| 2 | N2 * | Poterioochromonas sp. | 99% | AM981258.1 |
| 3 | N3 * | Scenedesmus sp. | 100% | KF569754.1 |
| 4 | N4 | Desmodesmus communis | 97% | KF864475.1 |
| 5 | N5 * | Scenedesmus sp. | 99% | FR865732.1 |
| 6 | N6 * | Chlorella emersonii | 99% | FR865661.1 |
| 7 | N7 * | Bracteacoccus sp. | 97% | JQ259919.1 |
| 8 | N8 * | Scenedesmus armatus | 100% | KR082490.1 |
| 9 | N9 * | Chlorella sorokiniana | 99% | KF444207.1 |
| 10 | N10 | Monoraphidium sp. | 100% | KR061995.1 |
| 11 | N11 | Selenastrum sp. | 99% | JQ360530.1 |
| 12 | N12 * | Acutodesmus sp. | 99% | KT267272.1 |
| 13 | N13 * | Chlorella sp. | 99% | FR865687.1 |
| 14 | N14 | Chlorella sp. | 99% | KT452082.1 |
| 15 | N15 | Ankistrodesmus gracilis | 94% | KF574394.1 |
| 16 | N16 * | Mychonastes sp. | 99% | JN617908.1 |
| 17 | N17 | Kirchneriella dianae | 99% | HM483512.1 |
| 18 | N18 * | Raphidocelis subcapitata | 100% | KF673369.1 |
| 19 | N19 | Dictyosphaerium sp. | 100% | GQ176860.1 |
| 20 | N20 | Desmodesmus abundans | 99% | AB917136.1 |
| 21 | N21 | Muriella decolor | 99% | JN968587.1 |
| 22 | N22 * | Scenedesmus sp. | 100% | KF569755.1 |
| 23 | N23 | Neodesmus sp. | 99% | KJ173793.1 |
| 24 | N24 * | Scenedesmus sp. | 100% | KT267272.1 |
| 25 | L1 | Chlorella sorokiniana | 100% | KR092112.1 |
| 26 | L2 * | Dictyosphaerium sp. | 99% | GQ487254.1 |
| 27 | L3 * | Coelastrella sp. | 99% | JX513883.1 |
| 28 | L6 | Schizochlamydella capsulata | 95% | AY044652.1 |
| 29 | L7 | Chlorella vulgaris | 99% | FR865683.1 |
| 30 | L8 * | Oocystidium sp. | 96% | HQ008711.1 |
| 31 | L9 | Nannochloris sp. | 100% | JQ315642.1 |
| 32 | L10 | Auxenochlorella protothecoides | 98% | KM462820.1 |
| 33 | L11 | Chlorella sp. | 99% | KP262476.1 |
| 34 | L12 * | Chlorella sorokiniana | 99% | KF864476.1 |
| 35 | L13 | Mychonastes afer | 99% | GQ477049.1 |
| 36 | M1 * | Nannochloris sp. | 98% | KF791551.1 |
| 37 | M2 | Nannochloris sp. | 100% | JQ315641.1 |
| 38 | M3 | Chlorosarcinopsis sp. | 98% | JN76.10864 |
| 39 | M4 | Stichococcus sp. | 99% | KM020184.1 |
| 40 | M5 | Picochlorum sp. | 100% | JN191236.1 |
| 41 | M6 | Chlorella sp. | 99% | EU282452.1 |
| 42 | M7 | Prasinoderma sp. | 99% | AB183584.1 |
| 43 | M8 | Picochlorum eukaryotum | 99% | X06425.1 |
| 44 | M9 | Chlorococcum sp. | 97% | AB183580.1 |
| 45 | M10 | Picochlorum maculatum | 99% | KM055115.1 |
| 46 | M11 * | Picochlorum oculatum | 99% | AY422075.1 |
| 47 | M12 | Marvania sp. | 98% | KF144207.1 |
| 48 | M13 | Chlorella minutissima | 99% | HQ218939.1 |
| 49 | M14 | Picochlorum atomus | 99% | FJ536747.1 |
| 50 | M15 | Nannochloris sp. | 99% | AB058309.1 |
| Strains | Saturated Fatty Acids (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C8:0 | C10:0 | C12:0 | C14:0 | C15:0 | C16:0 | C17:0 | C18:0 | C20:0 | C22:0 | C24:0 | Total | |
| Chlamydomonassp. N1 | 1.2 | 0.95 | 8.56 | 5.38 | 0.87 | 35.66 | 1.18 | 14.10 | 1.92 | 6.15 | - | 75.97 |
| Poterioochromonas sp. N2 | - | - | 0.38 | 1.35 | - | 33.72 | 0.54 | 10.56 | 1.53 | 3.44 | 4.27 | 55.79 |
| Scenedesmus sp. N3 | - | 0.17 | 1.48 | 1.55 | 0.24 | 34.09 | 0.51 | 8.57 | 1.07 | 3.84 | 1.43 | 52.95 |
| Scenedesmus sp. N5 | - | - | 0.87 | 0.70 | 0.17 | 18.13 | - | 1.87 | 0.09 | 0.24 | 0.22 | 22.29 |
| Chlorella emersonii N6 | - | 0.15 | 0.55 | 0.63 | 0.30 | 44.03 | 0.68 | 6.07 | 0.69 | 1.74 | 0.89 | 55.73 |
| Bracteacoccus sp. N7 | - | - | 0.47 | 2.21 | 0.37 | 25.91 | - | 7.83 | 1.66 | 4.25 | 4.05 | 46.75 |
| Scenedesmus armatus N8 | 1.88 | - | 1.99 | 1.79 | - | 34.72 | - | 15.00 | 1.17 | 4.77 | - | 61.32 |
| Chlorella sorokiniana N9 | - | - | - | 1.69 | - | 41.81 | 1.00 | 7.77 | 0.88 | 4.01 | 4.58 | 61.74 |
| Acutodesmus sp. N12 | - | - | - | - | - | 36.79 | 1.23 | 10.33 | 1.85 | 5.74 | 2.29 | 58.23 |
| Chlorella sp. N13 | - | - | - | 0.22 | 0.06 | 33.23 | 0.43 | 6.22 | 0.37 | 0.13 | - | 40.66 |
| Mychonastes sp. N16 | - | 0.30 | 3.14 | 2.27 | 0.56 | 51.54 | 0.91 | 8.97 | 1.17 | 2.87 | 6.56 | 78.05 |
| Raphidocelis subcapitata N18 | 2.29 | 1.92 | 5.07 | 3.34 | 0.75 | 40.54 | 0.95 | 12.93 | 1.31 | 3.93 | 3.26 | 76.29 |
| Scenedesmus sp. N22 | 0.38 | 0.47 | 2.00 | 3.06 | 0.47 | 47.85 | 0.74 | 10.38 | 0.77 | 2.23 | 1.04 | 69.39 |
| Scenedesmus sp. N24 | - | - | - | 0.66 | 0.24 | 25.22 | - | 2.19 | - | - | - | 28.31 |
| Dictyosphaerium sp. L2 | 0.55 | 0.48 | 3.88 | 2.76 | - | 31.46 | - | 5.09 | 0.53 | 1.74 | - | 46.49 |
| Coelastrellas p. L3 | 0.43 | 0.25 | 2.82 | 2.46 | 0.60 | 35.98 | 0.83 | 14.54 | 1.26 | 3.80 | - | 62.97 |
| Oocystidium sp. L8 | - | - | - | 0.97 | - | 18.53 | - | 3.60 | 0.31 | 2.39 | - | 25.80 |
| Chlorella sorokiniana L12 | - | - | - | 0.35 | - | 23.48 | 0.37 | 5.19 | 0.52 | 0.22 | - | 30.07 |
| Nannochloris sp. M1 | - | - | - | 0.63 | - | 27.08 | - | 1.41 | - | - | - | 29.12 |
| Picochlorum oculatum M11 | - | - | - | - | - | 29.60 | - | 2.35 | - | - | - | 31.95 |
| Strains | Unsaturated Fatty Acid (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C14:1 | C15:1 | C16:1 | C18:1 | C18:2 | C18:3 (α ) | C18:3 (γ) | C20:1 | C22:1 | MUFA | PUFA | Total | |
| Chlamydomonas sp. N1 | - | - | 1.83 | 14.12 | 2.86 | 2.59 | - | 0.56 | - | 16.51 | 5.45 | 21.96 |
| Poterioochromonas sp. N2 | - | - | 2.42 | 28.45 | 6.07 | 1.74 | - | 1.02 | 0.71 | 32.60 | 7.81 | 40.41 |
| Scenedesmus sp. N3 | - | - | - | 26.16 | 8.08 | 4.28 | - | 1.23 | 0.50 | 27.89 | 15.38 | 43.27 |
| Scenedesmus sp. N5 | 0.20 | - | 7.62 | 13.51 | 15.23 | 22.39 | -- | 0.15 | 0.20 | 21.68 | 38.30 | 59.98 |
| Chlorella emersonii N6 | - | - | 5.72 | 26.00 | 4.39 | 1.73 | - | 0.60 | - | 32.32 | 6.12 | 38.44 |
| Bracteacoccus sp. N7 | - | - | 2.59 | 26.51 | 11.14 | 3.25 | - | 1.21 | 0.75 | 31.06 | 14.39 | 45.45 |
| Scenedesmus armatus N8 | - | - | - | 30.72 | 6.29 | 1.07 | - | 0.59 | - | 31.31 | 7.36 | 38.67 |
| Chlorella sorokiniana N9 | - | - | 3.02 | 19.04 | 7.16 | 2.43 | - | 0.29 | - | 22.35 | 9.59 | 31.94 |
| Acutodesmus sp. N12 | - | - | 0.67 | 29.42 | 3.67 | 1.17 | - | - | - | 30.09 | 9.43 | 39.52 |
| Chlorella sp. N13 | 0.09 | - | 2.78 | 31.55 | 11.16 | 4.22 | 1.99 | 0.91 | 0.07 | 35.40 | 17.37 | 52.77 |
| Mychonastes sp. N16 | - | - | 1.61 | 14.59 | 2.24 | 0.74 | - | 1.17 | 0.78 | 18.15 | 2.98 | 21.13 |
| Raphidocelis subcapitata N18 | - | - | 1.53 | 11.65 | 3.37 | 2.31 | - | 0.81 | 0.84 | 14.83 | 5.68 | 20.51 |
| Scenedesmus sp. N22 | - | - | 0.54 | 15.48 | 5.85 | 4.14 | - | 0.92 | 0.45 | 17.39 | 9.99 | 27.38 |
| Scenedesmus sp. N24 | - | - | 2.11 | 13.82 | 8.70 | 20.36 | 0.41 | - | - | 15.93 | 35.47 | 51.40 |
| Dictyosphaerium sp. L2 | - | - | - | 8.24 | 17.08 | 13.37 | - | 0.47 | 0.30 | 9.98 | 30.45 | 40.43 |
| Coelastrella sp. L3 | - | - | 2.64 | 19.53 | 6.24 | 3.82 | 0.32 | 1.21 | 1.14 | 24.52 | 10.38 | 34.90 |
| Oocystidium sp. L8 | - | - | 0.59 | 6.83 | 17.22 | 18.52 | 0.56 | 0.13 | - | 7.55 | 39.82 | 47.37 |
| Chlorella sorokiniana L12 | - | - | 0.96 | 33.80 | 18.36 | 4.48 | 0.05 | 0.16 | - | 34.92 | 22.89 | 57.81 |
| Nannochloris sp. M1 | - | - | 1.44 | 7.30 | 20.04 | 10.91 | - | - | - | 8.74 | 30.95 | 39.69 |
| Picochlorum oculatum M11 | - | - | 0.51 | 6.50 | 22.29 | 17.20 | - | - | - | 7.01 | 39.49 | 46.50 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thao, T.Y.; Linh, D.T.N.; Si, V.C.; Carter, T.W.; Hill, R.T. Isolation and Selection of Microalgal Strains from Natural Water Sources in Viet Nam with Potential for Edible Oil Production. Mar. Drugs 2017, 15, 194. https://doi.org/10.3390/md15070194
Thao TY, Linh DTN, Si VC, Carter TW, Hill RT. Isolation and Selection of Microalgal Strains from Natural Water Sources in Viet Nam with Potential for Edible Oil Production. Marine Drugs. 2017; 15(7):194. https://doi.org/10.3390/md15070194
Chicago/Turabian StyleThao, Tran Yen, Dinh Thi Nhat Linh, Vo Chi Si, Taylor W. Carter, and Russell T. Hill. 2017. "Isolation and Selection of Microalgal Strains from Natural Water Sources in Viet Nam with Potential for Edible Oil Production" Marine Drugs 15, no. 7: 194. https://doi.org/10.3390/md15070194




