Molecular Identification of Gambierdiscus and Fukuyoa (Dinophyceae) from Environmental Samples
Abstract
:1. Introduction
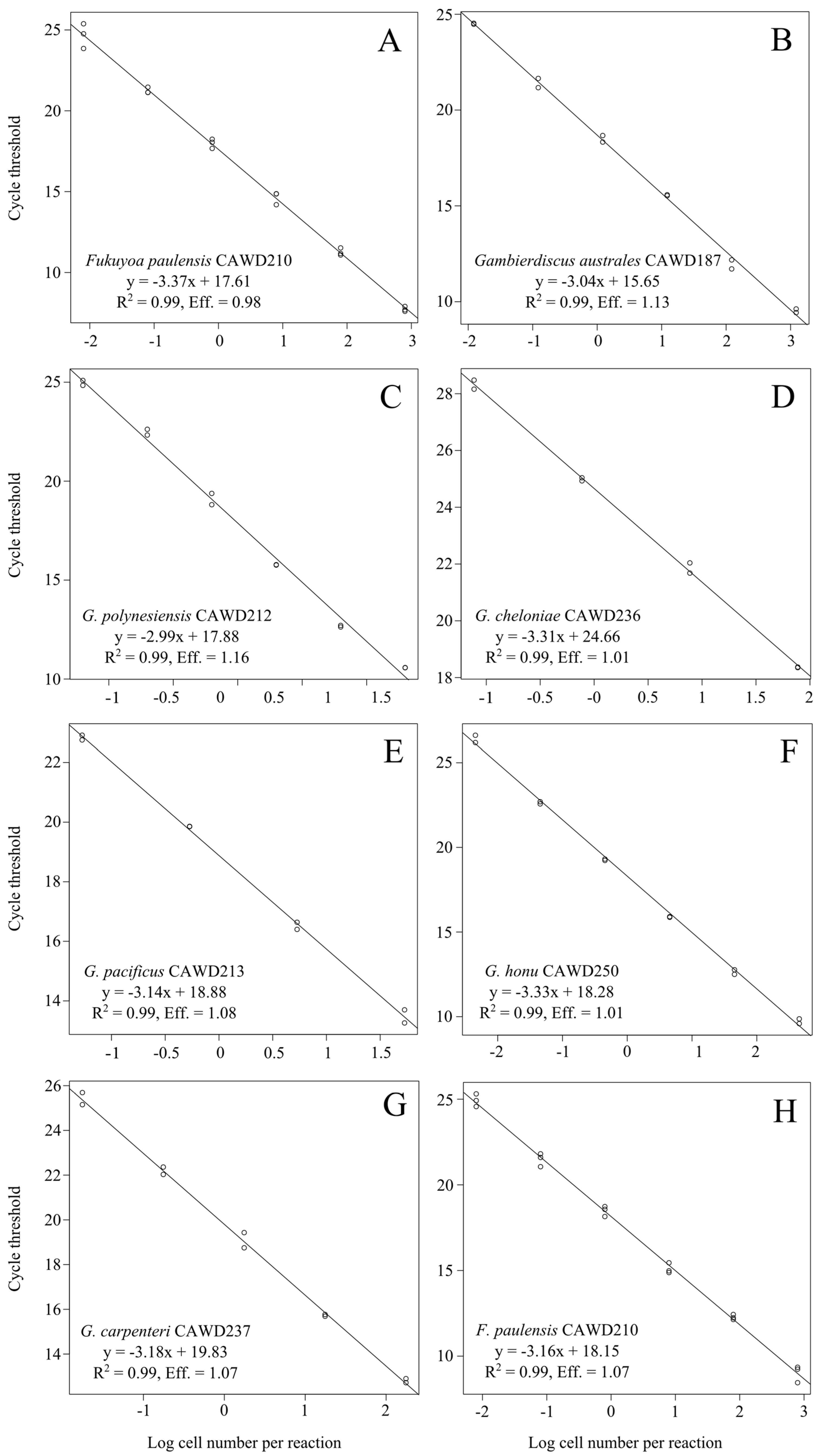
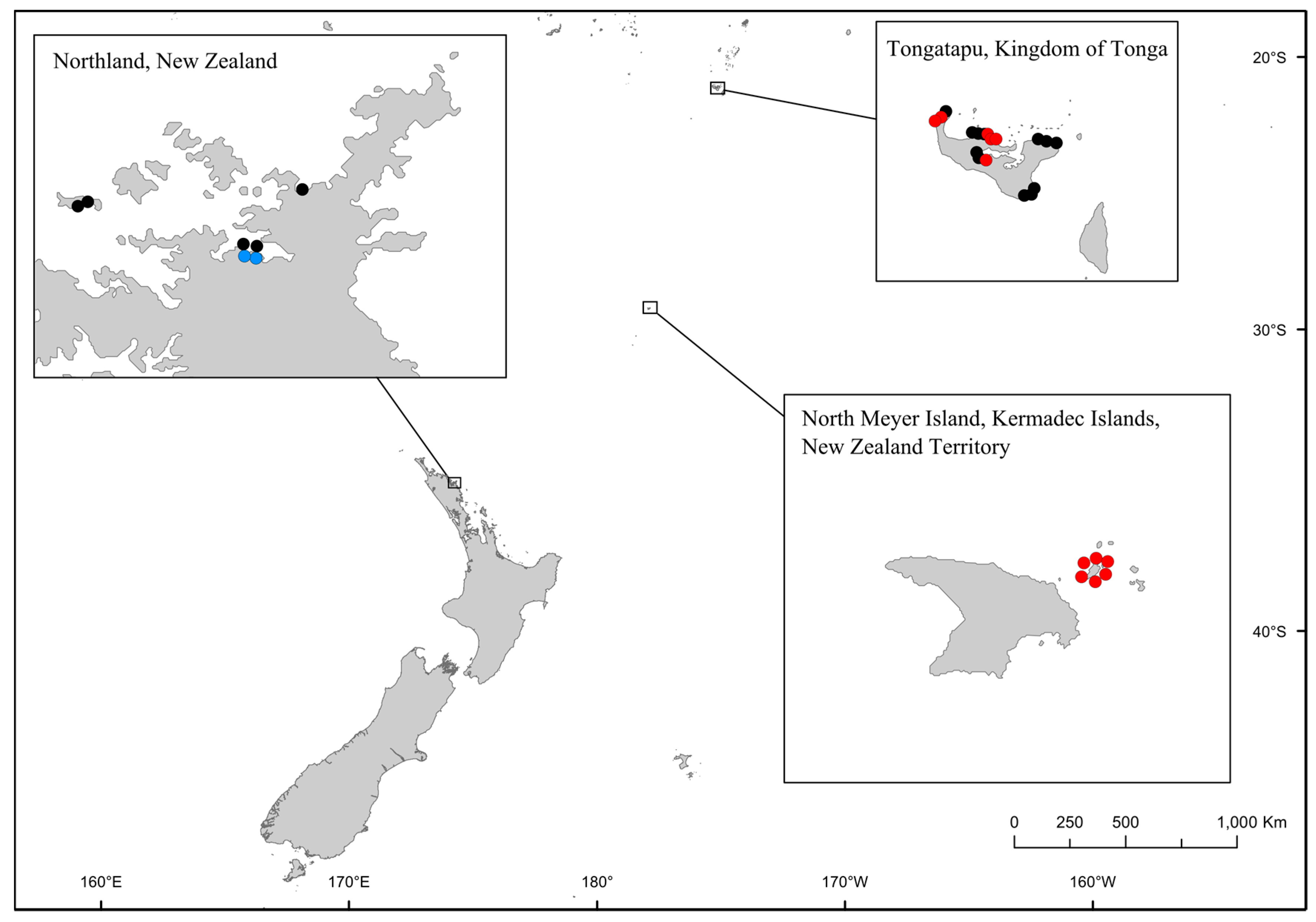
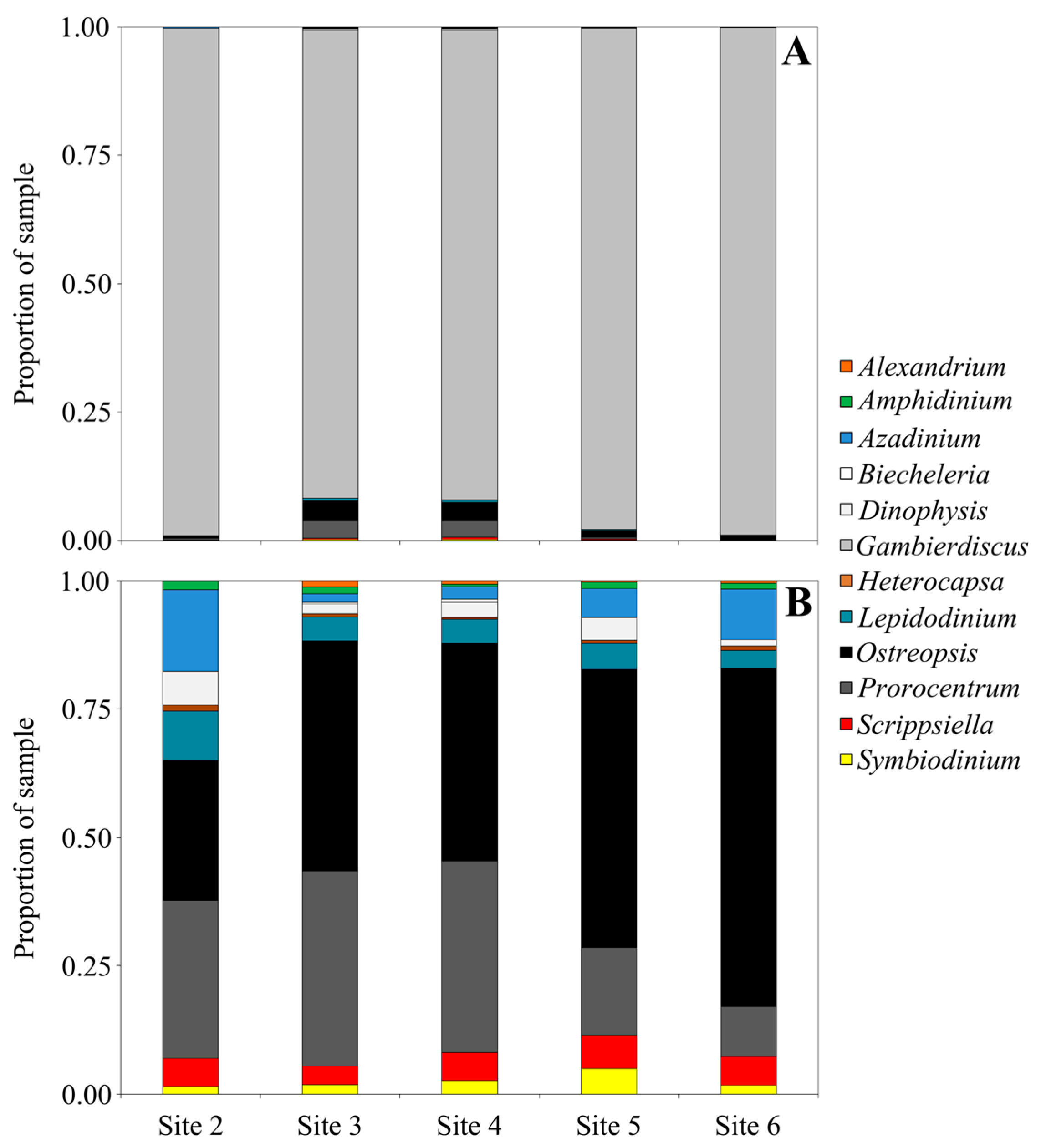
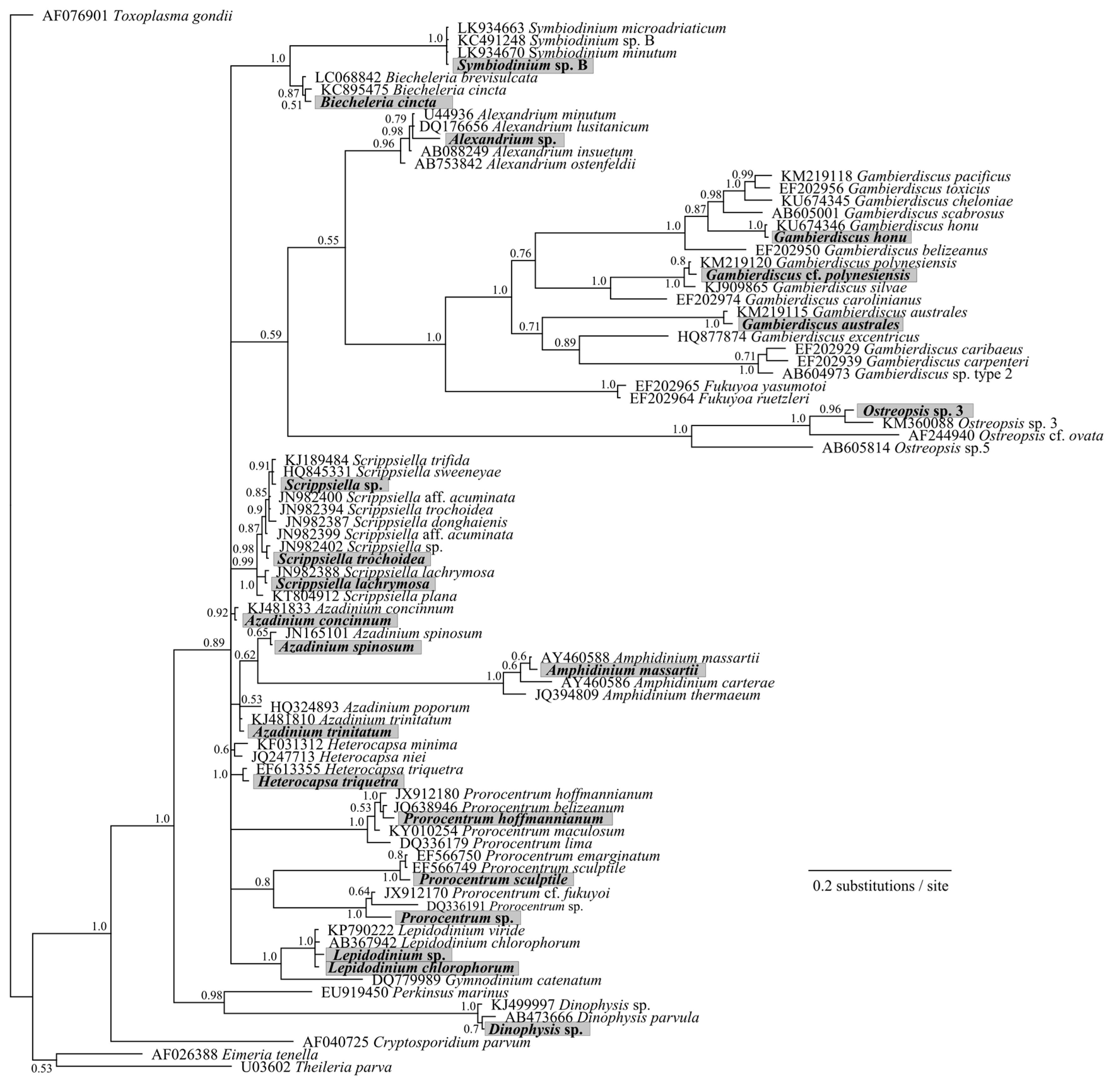
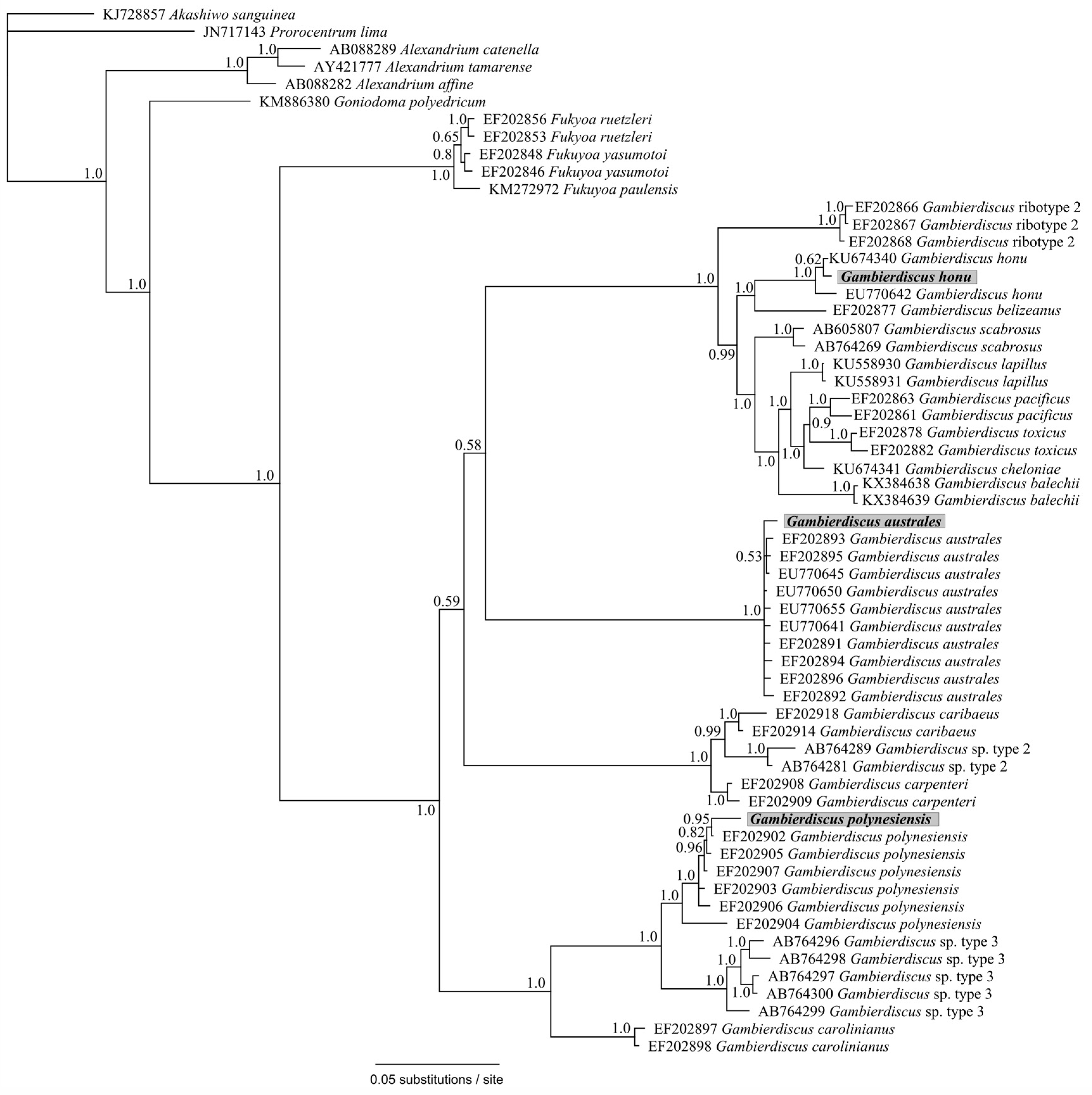
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Parsons, M.L.; Aligizaki, K.; Bottein, M.Y.D.; Fraga, S.; Morton, S.L.; Penna, A.; Rhodes, L. Gambierdiscus and Ostreopsis: Reassessment of the state of knowledge of their taxonomy, geography, ecophysiology, and toxicology. Harmful Algae 2012, 14, 107–129. [Google Scholar] [CrossRef]
- Hoppenrath, M.; Murray, S.A.; Chomerat, N.; Horiguchi, T. Marine Benthic Dinoflagellates—Unveiling Their Worldwide Biodiversity; Schweizerbart: Stuttgart, Germany, 2014. [Google Scholar]
- Kohli, G.S.; Farrell, H.; Murray, S.A. Gambierdiscus, the cause of ciguatera fish poisoning: An increased human health threat influenced by climate change. In Climate Change and Marine and Freshwater Toxins; Botana, L.M., Louzao, C., Vilarino, N., Eds.; De Gruyter: Berlin, Germany, 2015; pp. 273–312. [Google Scholar]
- Laza-Martínez, A.; David, H.; Riobó, P.; Miguel, I.; Orive, E. Characterization of a strain of Fukuyoa paulensis (Dinophyceae) from the Western Mediterranean Sea. J. Eukaryot. Microbiol. 2016, 63, 481–497. [Google Scholar] [CrossRef] [PubMed]
- Kohli, G.S.; Papiol, G.G.; Rhodes, L.L.; Harwood, D.T.; Selwood, A.; Jerrett, A.; Murray, S.A.; Neilan, B.A. A feeding study to probe the uptake of maitotoxin by snapper (Pagrus auratus). Harmful Algae 2014, 37, 125–132. [Google Scholar] [CrossRef]
- Berdalet, E.; Bravo, I.; Evans, J.; Fraga, S.; Kibler, S.; Kudela, M.; Larsen, J.; Litaker, W.; Penna, A.; Tester, P.; et al. Global Ecology and Oceanography of Harmful Algal Blooms, GEOHAB Core Research Project: HABs in Benthic Systems; Intergovernmental Oceanographic Commission: Lausanne, Switzerland, 2012. [Google Scholar]
- Adachi, R.; Fukuyo, Y. The thecal structure of a marine toxic dinoflagellate Gambierdiscus toxicus gen. et sp. nov. collected in a Ciguatera-endemic area. Bull. Jpn. Soc. Sci. Fish. 1979, 45, 67–71. [Google Scholar] [CrossRef]
- Bagnis, R.; Chanteau, S.; Chungue, E.; Hurtel, J.M.; Yasumoto, T.; Inoue, A. Origins of Ciguatera Fish Poisoning: A new dinoflagellate, Gambierdiscus toxicus Adachi and Fukuyo, definitively involved as a causal agent. Toxicon 1980, 18, 199–208. [Google Scholar] [CrossRef]
- Litaker, R.W.; Vandersea, M.W.; Faust, M.A.; Kibler, S.R.; Chinain, M.; Holmes, M.J.; Holland, W.C.; Tester, P.A. Taxonomy of Gambierdiscus including four new species, Gambierdiscus caribaeus, Gambierdiscus carolinianus, Gambierdiscus carpenteri and Gambierdiscus ruetzleri (Gonyaulacales, Dinophyceae). Phycologia 2009, 48, 344–390. [Google Scholar] [CrossRef]
- Rongo, T.; van Woesik, R. Socioeconomic consequences of Ciguatera poisoning in Rarotonga, Southern Cook Islands. Harmful Algae 2012, 20, 92–100. [Google Scholar] [CrossRef]
- Vandersea, M.W.; Kibler, S.R.; Holland, W.C.; Tester, P.A.; Schultz, T.F.; Faust, M.A.; Holmes, M.J.; Chinain, M.; Litaker, R.W. Development of semi-quantitative pcr assays for the detection and enumeration of Gambierdiscis species (Gonyaulacales, Dinophyceae). J. Phycol. 2012, 48, 902–915. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Hariganeya, N.; Tawong, W.; Sakanari, H.; Yamaguchi, H.; Adachi, M. Quantitative PCR assay for detection and enumeration of Ciguatera-causing dinoflagellate Gambierdiscus spp. (Gonyaulacales) in coastal areas of japan. Harmful Algae 2016, 52, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Faust, M.A. Observation of sand-dwelling toxic dinoflagellates (Dinophyceae) from widely differing sites, including two new species. J. Phycol. 1995, 31, 996–1003. [Google Scholar] [CrossRef]
- Chinain, M.; Faust, M.A.; Pauillac, S. Morphology and molecular analyses of three toxic species of Gambierdiscus (Dinophyceae): G. pacificus, sp. nov., G. australes, sp. nov., and G. polynesiensis, sp. nov. J. Phycol. 1999, 35, 1282–1296. [Google Scholar] [CrossRef]
- Fraga, S.; Rodríguez, F.; Caillaud, A.; Diogène, J.; Raho, N.; Zapata, M. Gambierdiscus excentricus sp. nov. (Dinophyceae), a benthic toxic dinoflagellate from the Canary Islands (NE Atlantic Ocean). Harmful Algae 2011, 11, 10–22. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, T.; Sato, S.; Tawong, W.; Sakanari, H.; Yamaguchi, H.; Adachi, M. Morphology of Gambierdiscus scabrosus sp. Nov. (Gonyaulacales): A new epiphytic toxic dinoflagellate from coastal areas of Japan. J. Phycol. 2014, 50, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Fraga, S.; Rodríguez, F. Genus Gambierdiscus in the Canary Islands (NE atlantic ocean) with description of Gambierdiscus silvae sp. nov., a new potentially toxic epiphytic benthic dinoflagellate. Protist 2014, 165, 839–853. [Google Scholar] [CrossRef] [PubMed]
- Fraga, S.; Rodríguez, F.; Riobó, P.; Bravo, I. Gambierdiscus balechii sp. nov (Dinophyceae), a new benthic toxic dinoflagellate from the Celebes Sea (SW Pacific Ocean). Harmful Algae 2016, 58, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.F.; Rhodes, L.; Verma, A.; Curley, B.G.; Harwood, D.T.; Kohli, G.S.; Solomona, D.; Rongo, T.; Munday, R.; Murray, S.A. A new Gambierdiscus species (Dinophyceae) from Rarotonga, Cook Islands: Gambierdiscus cheloniae sp. nov. Harmful Algae 2016, 60, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, L.; Smith, K.F.; Verma, A.; Curley, B.G.; Harwood, D.T.; Murray, S.; Kohli, G.S.; Solomona, D.; Rongo, T.; Munday, R.; et al. A new species of Gambierdiscus (Dinophyceae) from the South-West Pacific: Gambierdiscus honu sp. nov. Harmful Algae 2017, 65, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, A.L.; Verma, A.; Harwood, T.; Hoppenrath, M.; Murray, S. Characterization of Gambierdiscus lapillus sp. nov. (Gonyaulacales, Dinophyceae): A new toxic dinoflagellate from the Great Barrier Reef (Australia). J. Phycol. 2017, 53, 283–297. [Google Scholar] [CrossRef] [PubMed]
- Litaker, R.W.; Vandersea, M.W.; Faust, M.A.; Kibler, S.R.; Nau, A.W.; Holland, W.C.; Chinain, M.; Holmes, M.J.; Tester, P.A. Global distribution of Ciguatera causing dinoflagellates in the genus Gambierdiscus. Toxicon 2010, 56, 711–730. [Google Scholar] [CrossRef] [PubMed]
- Kuno, S.; Kamikawa, R.; Yoshimatsu, S.; Sagara, T.; Nishio, S.; Sako, Y. Genetic diversity of Gambierdiscus spp. (Gonyaulacales, Dinophyceae) in Japanese coastal areas. Phycol. Res. 2010, 58, 44–52. [Google Scholar] [CrossRef]
- Nishimura, T.; Sato, S.; Tawong, W.; Sakanari, H.; Uehara, K.; Shah, M.M.R.; Suda, S.; Yasumoto, T.; Taira, Y.; Yamaguchi, H.; et al. Genetic diversity and distribution of the Ciguatera-causing dinoflagellate Gambierdiscus spp. (Dinophyceae) in coastal areas of Japan. PLoS ONE 2013, 8, e60882. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Richlen, M.L.; Morton, S.L.; Mak, Y.L.; Chan, L.L.; Tekiau, A.; Anderson, D.M. Distribution, abundance and diversity of Gambierdiscus spp. from a Ciguatera-endemic area in Marakei, republic of Kiribati. Harmful Algae 2014, 34, 56–68. [Google Scholar] [CrossRef]
- Holmes, M.J. Gambierdiscus yasumotoi sp. nov. (Dinophyceae), a toxic benthic dinoflagellate from Southeastern Asia. J. Phycol. 1998, 34, 661–668. [Google Scholar] [CrossRef]
- Gómez, F.; Qiu, D.; Lopes, R.M.; Lin, S. Fukuyoa paulensis gen. et sp. nov., a new genus for the globular species of the dinoflagellate Gambierdiscus (Dinophyceae). PLoS ONE 2015, 10, e0119676. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, L.L.; Smith, K.F.; Verma, A.; Murray, S.; Harwood, D.T.; Trnski, T. The dinoflagellate genera Gambierdiscus and Ostreopsis from subtropical Raoul Island and North Meyer Island, Kermadec Islands. N. Z. J. Mar. Freshw. Res. 2017. [Google Scholar] [CrossRef]
- Smith, K.F.; Kohli, G.S.; Murray, S.A.; Rhodes, L.L. Assessment of the metabarcoding approach for community analysis of benthic-epiphytic dinoflagellates using mock communities. N. Z. J. Mar. Freshw. Res. 2017. [Google Scholar] [CrossRef]
- Kohli, G.S.; Neilan, B.A.; Brown, M.V.; Hoppenrath, M.; Murray, S.A. Cob gene pyrosequencing enables characterization of benthic dinoflagellate diversity and biogeography. Environ. Microbiol. 2013, 16, 467–485. [Google Scholar] [CrossRef] [PubMed]
- Le Bescot, N.; Mahé, F.; Audic, S.; Dimier, C.; Garet, M.-J.; Poulain, J.; Wincker, P.; Vargas, C.; Siano, R. Global patterns of pelagic dinoflagellate diversity across protist size classes unveiled by metabarcoding. Environ. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Bik, H.M.; Porazinska, D.L.; Creer, S.; Caporaso, J.G.; Knight, R.; Thomas, W.K. Sequencing our way towards understanding global eukaryotic biodiversity. Trends Ecol. Evol. 2012, 27, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Lallias, D.; Hiddink, J.G.; Fonseca, V.G.; Gaspar, J.M.; Sung, W.; Neill, S.P.; Barnes, N.; Ferrero, T.; Hall, N.; Lambshead, P.J.D.; et al. Environmental metabarcoding reveals heterogeneous drivers of microbial eukaryote diversity in contrasting estuarine ecosystems. ISME J. 2015, 9, 1208–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massana, R.; Gobet, A.; Audic, S.; Bass, D.; Bittner, L.; Boutte, C.; Chambouvet, A.; Christen, R.; Claverie, J.-M.; Decelle, J.; et al. Marine protist diversity in European coastal waters and sediments as revealed by high-throughput sequencing. Environ. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skinner, M.P.; Brewer, T.D.; Johnstone, R.; Fleming, L.E.; Lewis, R.J. Ciguatera fish poisoning in the Pacific Islands (1998 to 2008). PLoS Negl. Trop. Dis. 2011, 5, e1416. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, L.; Papiol, G.G.; Smith, K.; Harwood, T. Gambierdiscus cf. yasumotoi (Dinophyceae) isolated from new zealand’s sub-tropical northern coastal waters. N. Z. J. Mar. Freshw. Res. 2014, 48, 303–310. [Google Scholar] [CrossRef]
- Kohli, G.S.; Murray, S.A.; Neilan, B.A.; Rhodes, L.L.; Harwood, D.T.; Smith, K.F.; Meyer, L.; Capper, A.; Brett, S.; Hallegraeff, G.M. High abundance of the potentially maitotoxic dinoflagellate Gambierdiscus carpenteri in temperate waters of new south wales, australia. Harmful Algae 2014, 39, 134–145. [Google Scholar] [CrossRef]
- Chang, F.H. Shellfish toxin update. Seaf. N. Z. 1996, 26. [Google Scholar]
- Rhodes, L.; Smith, K.; Papiol, G.G.; Adamson, J.; Harwood, T.; Munday, R. Epiphytic dinoflagellates in sub-tropical New Zealand, in particular the genus Coolia Meunier. Harmful Algae 2014, 34, 36–41. [Google Scholar] [CrossRef]
- Rhodes, L.; Smith, K.; Harwood, T.; Bedford, C. Novel and toxin-producing epiphytic dinoflagellates isolated from sub-tropical Raoul Island, Kermadec Islands group. N. Z. J. Mar. Freshw. Res. 2014, 48, 594–599. [Google Scholar] [CrossRef]
- Selwood, A.I.; van Ginkel, R.; Harwood, D.T.; McNabb, P.S.; Rhodes, L.R.; Holland, P.T. A sensitive assay for palytoxins, ovatoxins and ostreocins using LC-MS/MS analysis of cleavage fragments from micro-scale oxidation. Toxicon 2012, 60, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- De Wit, P.; Pespeni, M.H.; Ladner, J.T.; Barshis, D.J.; Seneca, F.; Jaris, H.; Therkildsen, N.O.; Morikawa, M.; Palumbi, S.R. The simple fool's guide to population genomics via RNA-seq: An introduction to high-throughput sequencing data analysis. Mol. Ecol. Resour. 2012, 12, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.; Peterson, D.; Biggs, P. SolexaQA: At-a-glance quality assessment of illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing MOTHUR: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Nylander, J.A.A. Mrmodeltest V2 Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Rhodes, L.L.; Smith, K.F.; Murray, S.; Harwood, D.T.; Trnski, T.; Munday, M. The epiphytic genus Gambierdiscus (Dinophyceae) in the Kermadec Islands and Zealandia regions of the southwestern Pacific and the associated risk of ciguatera fish poisoning. Mar. Drugs 2017, 15, 219. [Google Scholar] [CrossRef] [PubMed]
| Target | Sequence | Gene Region | Product Size | Type |
|---|---|---|---|---|
| Gambierdiscus/Fukuyoa | ||||
| GA668-F | 5′-CTGTRTGACCCGTCTTGAAAC-3′ | D2-D3 LSU | 208 bp | QPCR |
| GA875-R | 5′-GTTTCCCCTGVCTTGRCC-3′ | |||
| Fukuyoa paulensis | ||||
| FP128-F | 5′-AAAGTGGGAAAAGGGTGG-3′ | ITS | 128 bp | QPCR |
| FP253-R | 5′-TGTCACCGCACCAAAATC-3′ | |||
| Dinoflagellates | ||||
| D1R | 5′-ACCCGCTGAATTTAAGCATA-3′ | D1-D2 LSU | 365 bp | HTS Metabarcoding |
| 305R | 5′-TTTAAYTCTCTTTYCAAAGTCC-3′ | |||
| Dinoflagellates | ||||
| SSU556F | 5′-CGCGGTAATTCCAGCTYC-3′ | V4 SSU | 346 bp | HTS Metabarcoding |
| SSU911R | 5′-ATYCAAGAATTTCACCTCTGAC-3′ | |||
| Species | Strain | Gambierdiscus/Fukuyoa | Fukuyoa paulensis | ||
|---|---|---|---|---|---|
| Result | Melt Temp. | Result | Melt Temp. | ||
| Gambierdiscus pacificus | CAWD213 | +/+ | 86.0 | −/− | NA |
| Gambierdiscus australes | CAWD149 | +/+ | 86.0 | −/− | NA |
| Gambierdiscus polynesiensis | CAWD212 | +/+ | 85.5 | −/− | NA |
| Gambierdiscus cheloniae | CAWD232 | +/+ | 85.8 | −/− | NA |
| Gambierdiscus honu | CAWD233 | +/+ | 85.5 | −/− | NA |
| Gambierdiscus ribotype II | CCMP1655 | +/+ | 83.5 | −/− | NA |
| Gambierdiscus caribaeus | CCMP1733 | +/+ | 85.5 | −/− | NA |
| Gambierdiscus carpenteri | CCMP1654 | +/+ | 84.5 | −/− | NA |
| Gambierdiscus belizeanus | CCMP401 | +/+ | 85.7 | −/− | NA |
| Gambierdiscus carolinianus | Kenny6 | +/+ | 87.8 | −/− | NA |
| Fukuyoa ruetzleri | Gam1 | +/+ | 84.0 | −/− | NA |
| Fukuyoa paulensis | CAWD210 | +/+ | 83.3 | +/+ | 82.0 |
| Fukuyoa paulensis | CAWD211 | NT | NT | +/+ | 81.8 |
| Ostreopsis sp. | CAWD239 | −/− | NA | −/− | NA |
| Ostreopsis sp. | L138 | −/− | NA | −/− | NA |
| Coolia malayensis | L130 | −/− | NA | −/− | NA |
| Alexandrium margalefii | K53 | −/− | NA | −/− | NA |
| Alexandrium catenella | K54 | −/− | NA | −/− | NA |
| Prorocentrum cf. micans | L144 | −/− | NA | −/− | NA |
| Karenia mikimotoi | CAWD63 | −/− | NA | −/− | NA |
| Gymnodinium impudicum | CAWD139 | −/− | NA | −/− | NA |
| Amphidinium massartii | L132 | −/− | NA | −/− | NA |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, K.F.; Biessy, L.; Argyle, P.A.; Trnski, T.; Halafihi, T.; Rhodes, L.L. Molecular Identification of Gambierdiscus and Fukuyoa (Dinophyceae) from Environmental Samples. Mar. Drugs 2017, 15, 243. https://doi.org/10.3390/md15080243
Smith KF, Biessy L, Argyle PA, Trnski T, Halafihi T, Rhodes LL. Molecular Identification of Gambierdiscus and Fukuyoa (Dinophyceae) from Environmental Samples. Marine Drugs. 2017; 15(8):243. https://doi.org/10.3390/md15080243
Chicago/Turabian StyleSmith, Kirsty F., Laura Biessy, Phoebe A. Argyle, Tom Trnski, Tuikolongahau Halafihi, and Lesley L. Rhodes. 2017. "Molecular Identification of Gambierdiscus and Fukuyoa (Dinophyceae) from Environmental Samples" Marine Drugs 15, no. 8: 243. https://doi.org/10.3390/md15080243




