Purification and Characterization of a Novel Alginate Lyase from the Marine Bacterium Bacillus sp. Alg07
Abstract
:1. Introduction
2. Results and Discussion
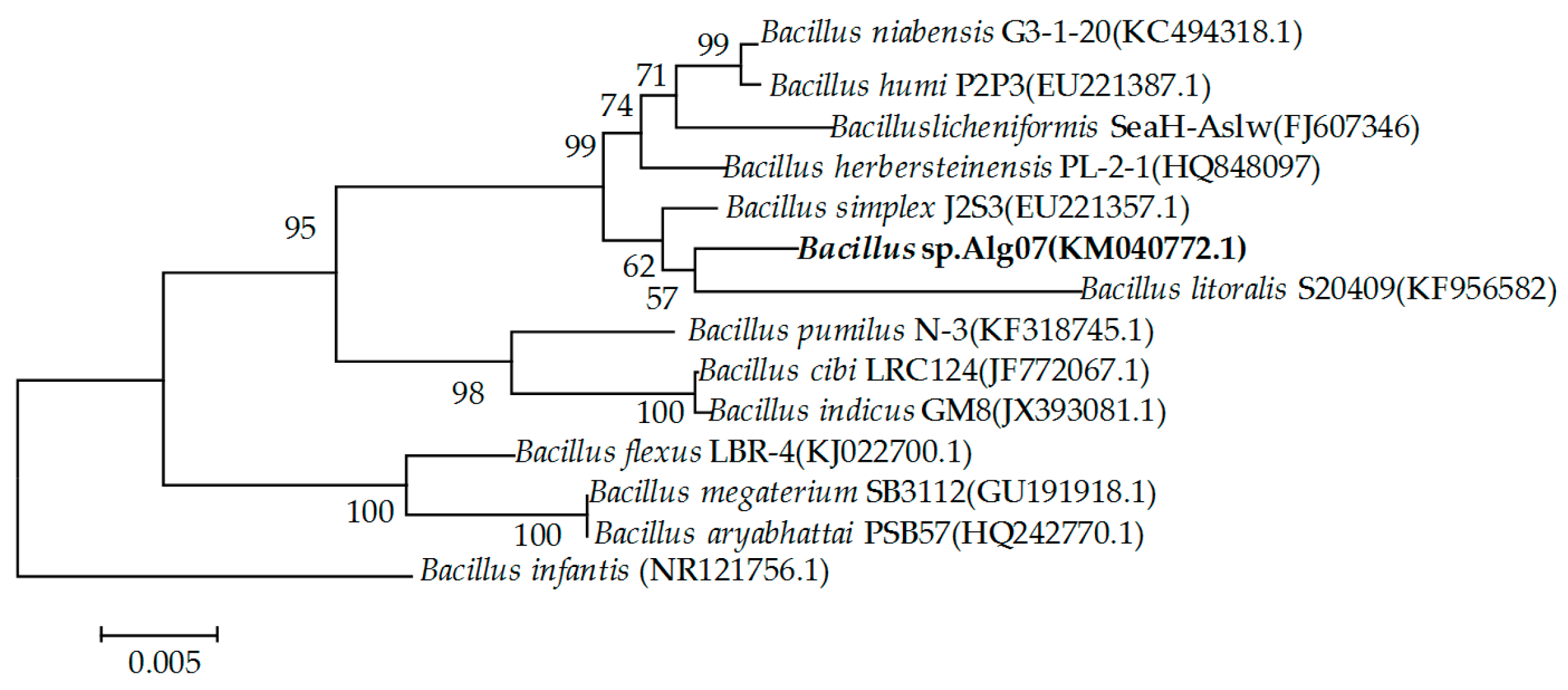
2.1. Screening and Identification of Strain Alg07
2.2. Purification of Alginate Lyase from Bacillus sp. Alg07
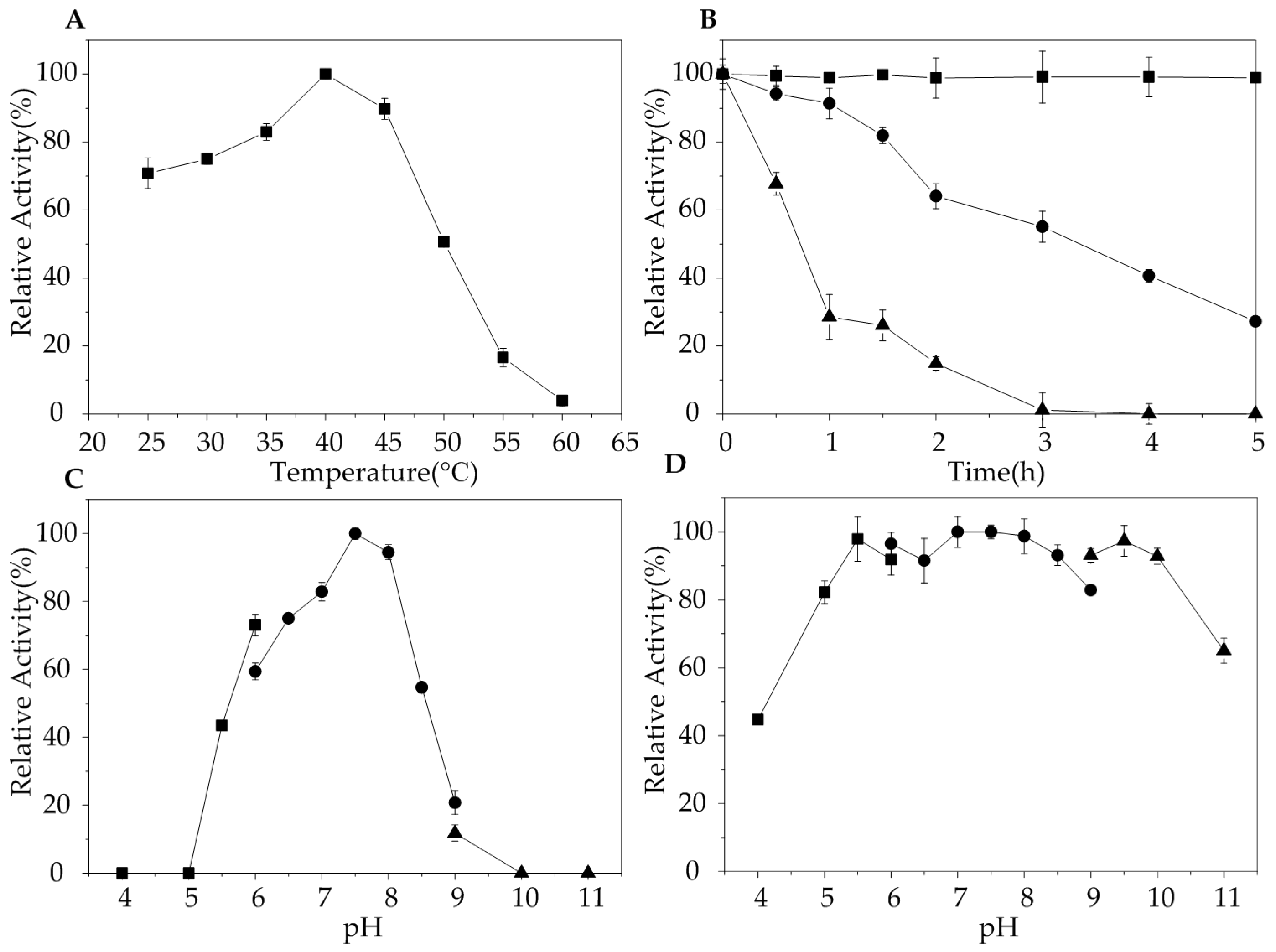
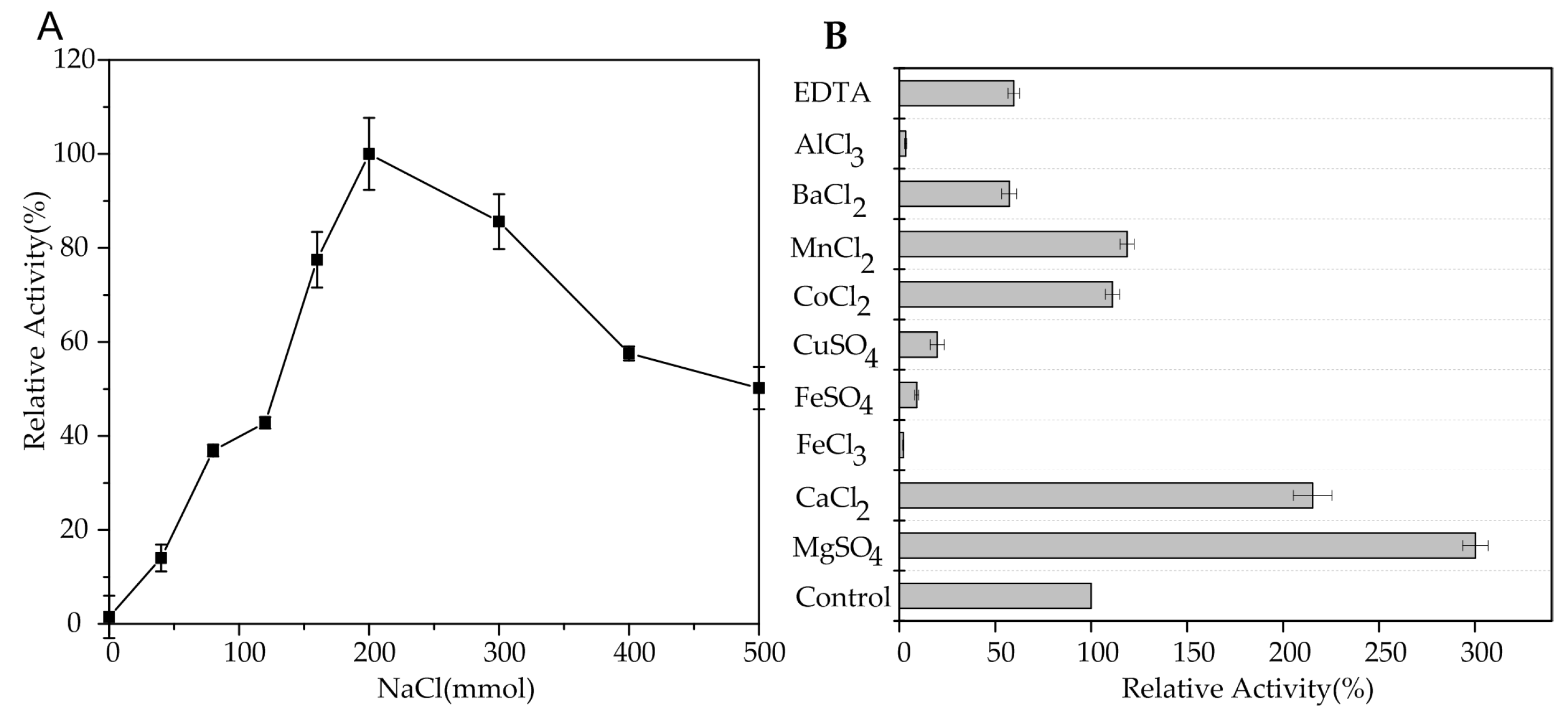
2.3. Biochemical Characterization of AlgA
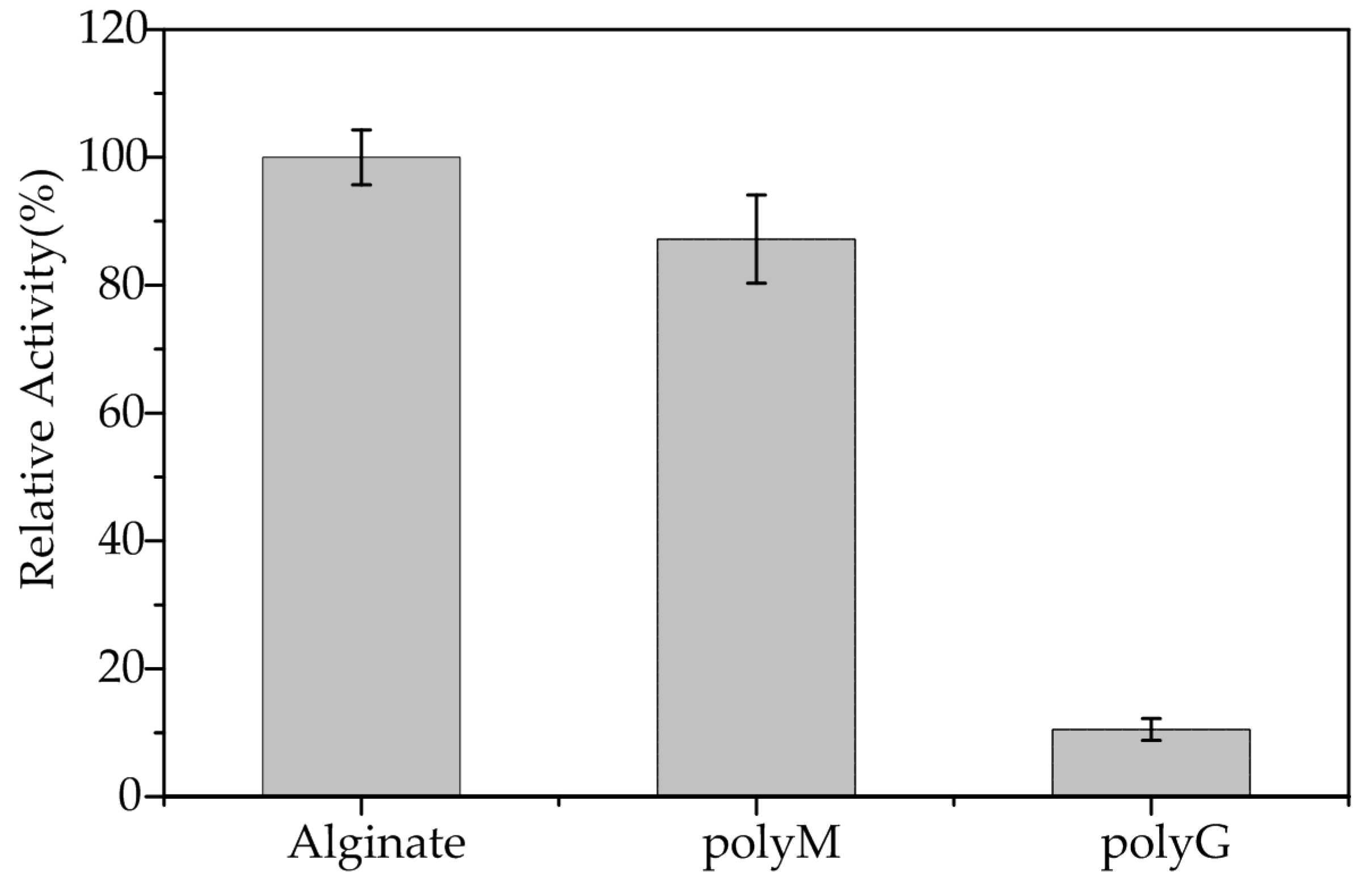
2.4. Substrate Specificity and Kinetic Parameters of AlgA
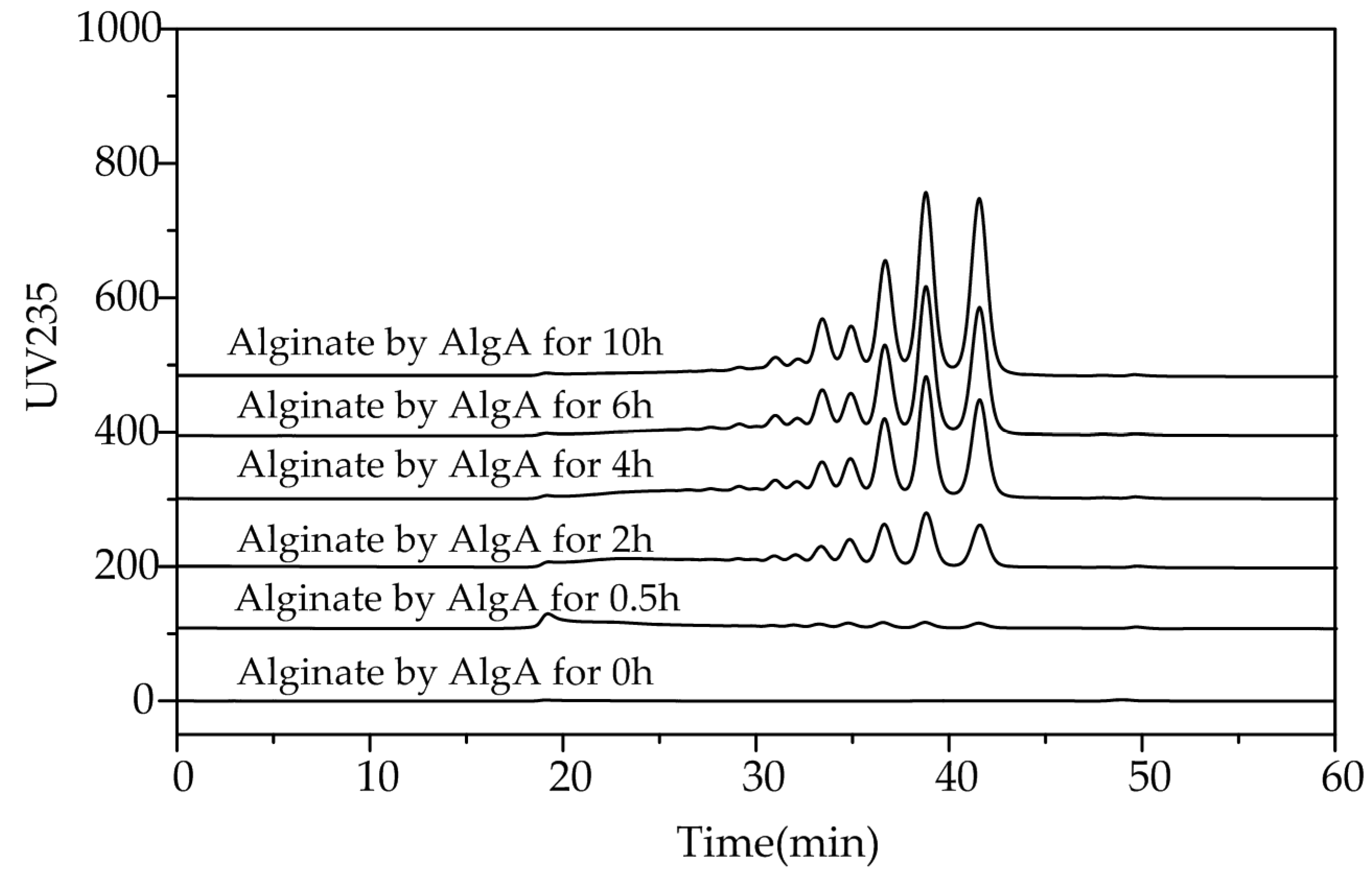
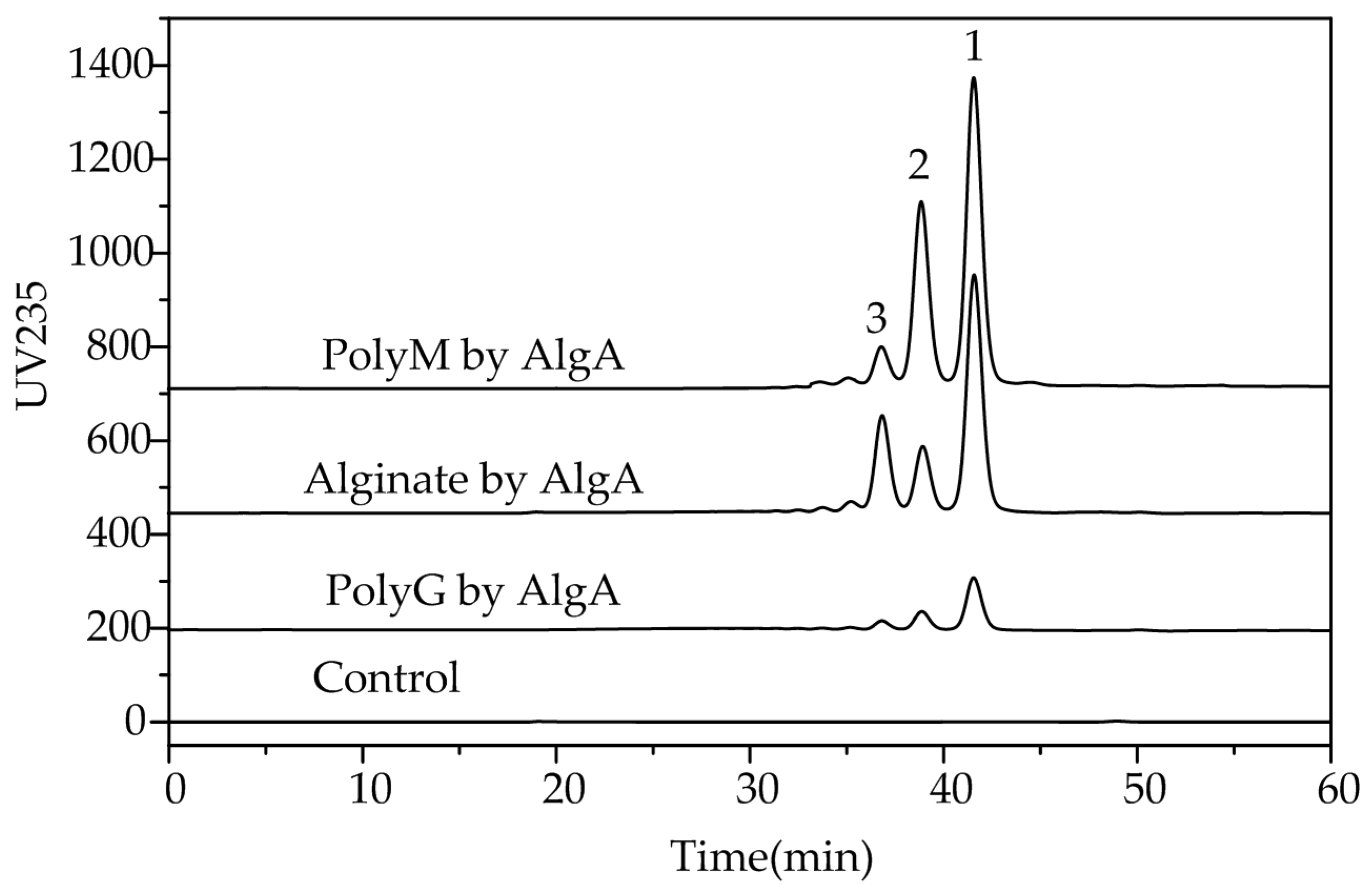
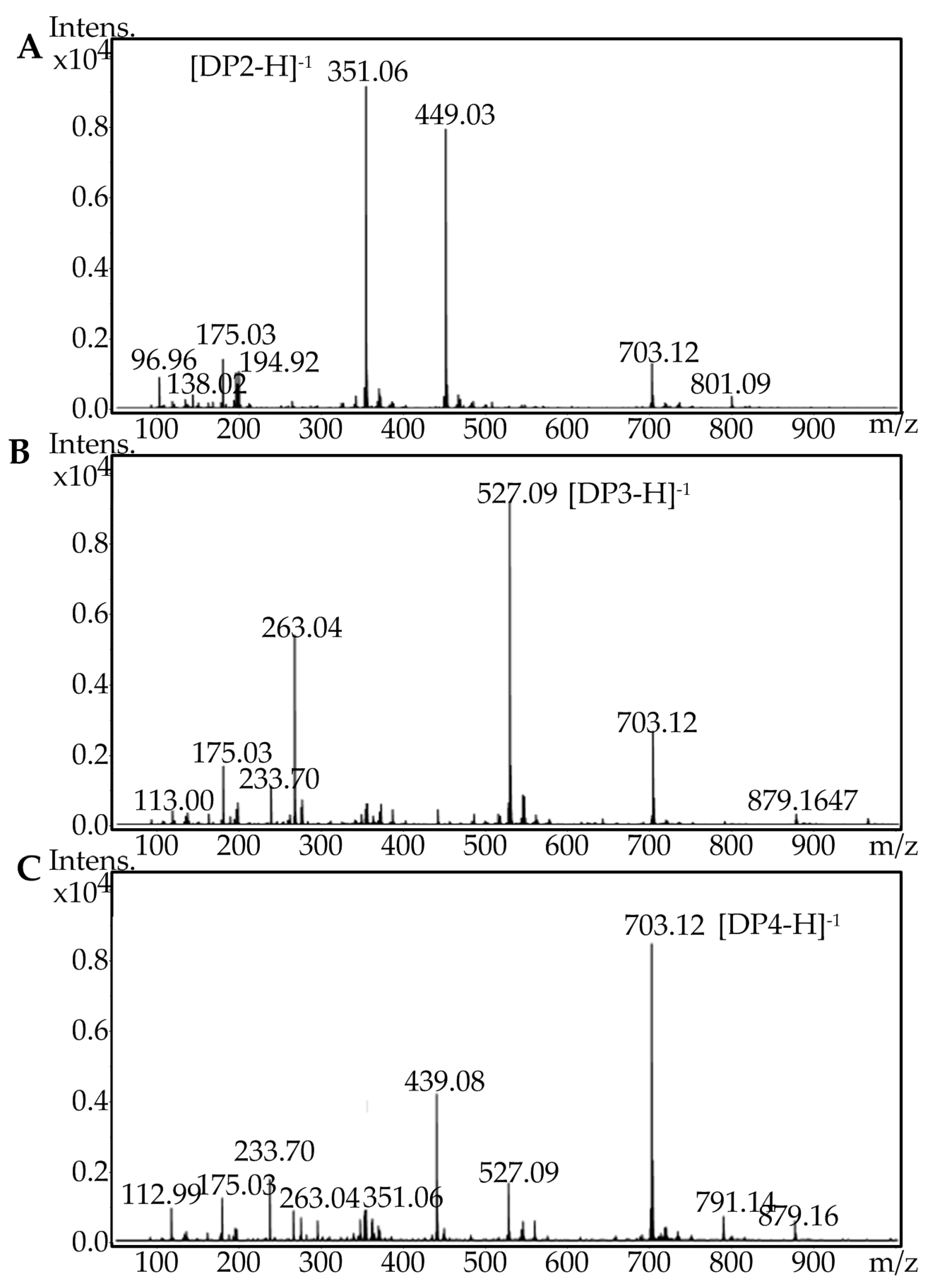
2.5. Fast Protein Liquid Chromatography and Electrospray Ionization Mass Spectrometry Analysis of the Degradation Products of AlgA
3. Materials and Methods
3.1. Materials
3.2. Screening and Identification of Strain Alg07
3.3. Production and Purification of AlgA
3.4. Enzyme Activity Assay
3.5. Characterization of AlgA
3.6. Analysis of the N-Terminal Amino Acid Sequence of AlgA
3.7. Substrate Specificity and Kinetic Parameters of AlgA
3.8. FPLC and ESI-MS Analysis of the Degradation Products of AlgA
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gacesa, P. Enzymic degradation of alginates. Int. J. Biochem. 1992, 24, 545–552. [Google Scholar] [CrossRef]
- Mabeau, S.; Kloareg, B. Isolation and Analysis of the Cell Walls of Brown Algae: Fucus spiralis, F. ceranoides, F. vesiculosus, F. serratus, Bifurcaria bifurcata and Laminaria digitata. J. Exp. Bot. 1987, 38, 1573–1580. [Google Scholar] [CrossRef]
- Gorin, P.A.J.; Spencer, J.F.T. Exocellular alginic acid from Azotobacter vinelandii. Can. J. Chem. 1966, 44, 993–998. [Google Scholar] [CrossRef]
- Evans, L.R.; Linker, A. Production and Characterization of the Slime Polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 1973, 116, 915–924. [Google Scholar] [PubMed]
- Scott, C.D. Immobilized cells: A review of recent literature. Enzym. Microb. Technol. 1987, 9, 66–72. [Google Scholar] [CrossRef]
- Kawada, A.; Hiura, N.; Tajima, S.; Takahara, H. Alginate oligosaccharides stimulate VEGF-mediated growth and migration of human endothelial cells. Arch. Dermatol. Res. 1999, 291, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kurachi, M.; Yamaguchi, K.; Oda, T. Stimulation of multiple cytokine production in mice by alginate oligosaccharides following intraperitoneal administration. Carbohydr. Res. 2007, 342, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, K.I.; Matsubara, Y. Purification of Alginate Oligosaccharides with Root Growth-promoting Activity toward Lettuce. Biosci. Biotechnol. Biochem. 2000, 64, 1067–1070. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, F.; Hu, B.; Li, J.; Yu, W. In vivo prebiotic properties of alginate oligosaccharides prepared through enzymatic hydrolysis of alginate. Nutr. Res. 2006, 26, 597–603. [Google Scholar] [CrossRef]
- Falkeborg, M.; Cheong, L.Z.; Gianfico, C.; Sztukiel, K.M.; Kristensen, K.; Glasius, M.; Xu, X.; Guo, Z. Alginate oligosaccharides: enzymatic preparation and antioxidant property evaluation. Food Chem. 2014, 164, 185–194. [Google Scholar] [CrossRef] [PubMed]
- An, Q.D.; Zhang, G.L.; Wu, H.T.; Zhang, Z.C.; Zheng, G.S.; Luan, L.; Murata, Y.; Li, X. Alginate-deriving oligosaccharide production by alginase from newly isolated Flavobacterium sp. LXA and its potential application in protection against pathogens. J. Appl. Microbiol. 2009, 106, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, C.G.; Lee, E.Y. Alginate lyase: Structure, property, and application. Biotechnol. Bioprocess Eng. 2011, 16, 843–851. [Google Scholar] [CrossRef]
- Inoue, A.; Mashino, C.; Uji, T.; Saga, N.; Mikami, K.; Ojima, T. Characterization of an Eukaryotic PL-7 Alginate Lyase in the Marine Red AlgaPyropia yezoensis. Curr. Biotechnol. 2015, 4, 240–258. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Suzuki, K.; Inoue, A.; Ojima, T. A novel oligoalginate lyase from abalone, Haliotis discus hannai, that releases disaccharide from alginate polymer in an exolytic manner. Carbohydr. Res. 2006, 341, 1809–1819. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Chen, M.; Yin, H.; Du, Y.; Ning, L. Enzymatic Hydrolysis of Alginate to Produce Oligosaccharides by a New Purified Endo-Type Alginate Lyase. Mar. Drugs. 2016, 14, 108. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Takadono, K.; Nishiyama, R.; Tajima, K.; Kobayashi, T.; Ojima, T. Characterization of an Alginate Lyase, FlAlyA, from Flavobacterium sp. Strain UMI-01 and Its Expression in Escherichia coli. Mar. Drugs 2014, 12, 4693–4712. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Gupta, V.; Kumari, P.; Kumar, M.; Reddy, C.R.K.; Prasad, K.; Jha, B. Purification and partial characterization of an extracellular alginate lyase from Aspergillus oryzae isolated from brown seaweed. J. Appl. Phycol. 2011, 23, 755–762. [Google Scholar] [CrossRef]
- Suda, K.; Tanji, Y.; Hori, K.; Unno, H. Evidence for a novel Chlorella virus-encoded alginate lyase. FEMS Microbiol. Lett. 1999, 180, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.Y.; Preston, L.A.; Schiller, N.L. ALGINATE LYASE: Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000, 54, 289–340. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Aarstad, O.A.; Tøndervik, A.; Sletta, H.; Skjåkbræk, G. Alginate Sequencing: An Analysis of Block Distribution in Alginates Using Specific Alginate Degrading Enzymes. Biomacromolecules 2012, 13, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Mashino, C.; Kodama, T.; Ojima, T. Protoplast preparation from Laminaria japonica with recombinant alginate lyase and cellulase. Mar. Biotechnol. 2011, 13, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Islan, G.A.; Martinez, Y.N.; Illanes, A.; Castro, G.R. Development of novel alginate lyase cross-linked aggregates for the oral treatment of cystic fibrosis. RSC Adv. 2014, 4, 11758–11765. [Google Scholar] [CrossRef]
- Li, L.; Jiang, X.; Guan, H.; Wang, P. Preparation, purification and characterization of alginate oligosaccharides degraded by alginate lyase from Pseudomonas sp. HZJ 216. Carbohydr. Res. 2011, 346, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, S.; Yu, W.; Gong, Q. Cloning, overexpression and characterization of a new oligoalginate lyase from a marine bacterium, Shewanella sp. Biotechnol. Lett. 2015, 37, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Ko, H.J.; Kim, N.; Kim, D.; Lee, D.; Choi, I.G.; Woo, H.C.; Kim, M.D.; Kim, K.H. Characterization of a recombinant endo-type alginate lyase (Alg7D) from Saccharophagus degradans. Biotechnol. Lett. 2012, 34, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, L.; Hao, J.; Xing, M.; Sun, J.; Sun, M. Purification and Characterization of a New Alginate Lyase from Marine Bacterium Vibrio sp. SY08. Mar. Drugs 2017, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, E.W.; Yu, W.G.; Han, F. Purification and characterization of a new alginate lyase from a marine bacterium Vibrio sp. Biotechnol. Lett. 2013, 35, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Uchimura, K.; Miyazaki, M.; Nogi, Y.; Horikoshi, K. A new high-alkaline alginate lyase from a deep-sea bacterium Agarivorans sp. Extremophiles 2009, 13, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.F.; Hong, L.; Sang, M.K. Purification and characterization of a Na+/K+ dependent alginate lyase from turban shell gut Vibrio sp. YKW-34. Enzym. Microb. Technol. 2007, 41, 828–834. [Google Scholar]
- Zhu, B.; Yin, H. Alginate lyase: Review of major sources and classification, properties, structure-function analysis and applications. Bioengineered 2015, 6, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, M.; Moriwaki, S.; Miyake, O.; Hashimoto, W.; Murata, K.; Mikami, B. Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7. J. Biol. Chem. 2004, 279, 31863–31872. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.I.; Choi, S.H.; Lee, E.Y.; Kim, H.S. Molecular cloning, purification, and characterization of a novel polyMG-specific alginate lyase responsible for alginate MG block degradation in Stenotrophomas maltophilia KJ-2. Appl. Microbiol. Biotechnol. 2012, 95, 1643–1653. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Tan, H.; Qin, Y.; Xu, Q.; Du, Y.; Yin, H. Characterization of a new endo-type alginate lyase from Vibrio sp. W13. Int. J. Biol. Macromol. 2015, 75, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.; Lundqvist, L.C.E.; Jam, M.; Jeudy, A.; Barbeyron, T.; Sandström, C.; Michel, G.; Czjzek, M. Comparative Characterization of Two Marine Alginate Lyases from Zobellia galactanivorans Reveals Distinct Modes of Action and Exquisite Adaptation to Their Natural Substrate. J. Biol. Chem. 2013, 288, 23021–23037. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, X.; Bao, M.; Wu, Y.; Yu, W.; Han, F. Family 13 carbohydrate-binding module of alginate lyase from Agarivorans sp. L11 enhances its catalytic efficiency and thermostability, and alters its substrate preference and product distribution. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Gu, J.; Cheng, Y.; Liu, H.; Li, Y.; Li, F. Novel Alginate Lyase (Aly5) from a Polysaccharide-Degrading Marine Bacterium, Flammeovirga sp. Strain MY04: Effects of Module Truncation on Biochemical Characteristics, Alginate Degradation Patterns, and Oligosaccharide-Yielding Properties. Appl. Environ. Microbiol. 2015, 82, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.; Dong, S.; Song, J.; Li, C.B.; Chen, X.L.; Xie, B.B.; Zhang, Y.Z. Purification and Characterization of a Bifunctional Alginate Lyase from Pseudoalteromonas sp. SM0524. Mar. Drugs 2011, 9, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Uchimura, K.; Miyazaki, M.; Nogi, Y.; Kobayashi, T.; Horikoshi, K. Cloning and sequencing of alginate lyase genes from deep-sea strains of Vibrio and Agarivorans and characterization of a new Vibrio enzyme. Mar. Biotechnol. 2010, 12, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Han, F.; Yang, Z.; Lu, X.Z.; Yu, W.G. A Novel Alginate Lyase with High Activity on Acetylated Alginate of Pseudomonas aeruginosa FRD1 from Pseudomonas sp. QD03. World J. Microbiol. Biotechnol. 2006, 22, 81–88. [Google Scholar] [CrossRef]
- Ueno, M.; Hiroki, T.; Takeshita, S.; Jiang, Z.; Kim, D.; Yamaguchi, K.; Oda, T. Comparative study on antioxidative and macrophage-stimulating activities of polyguluronic acid (PG) and polymannuronic acid (PM) prepared from alginate. Carbohydr. Res. 2012, 352, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Biochem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Haug, A.; Larsen, B. A study on the constitution of alginic acid by partial acid hydrolysis. Acta Chem. Scand. 1966, 20, 183–190. [Google Scholar] [CrossRef]
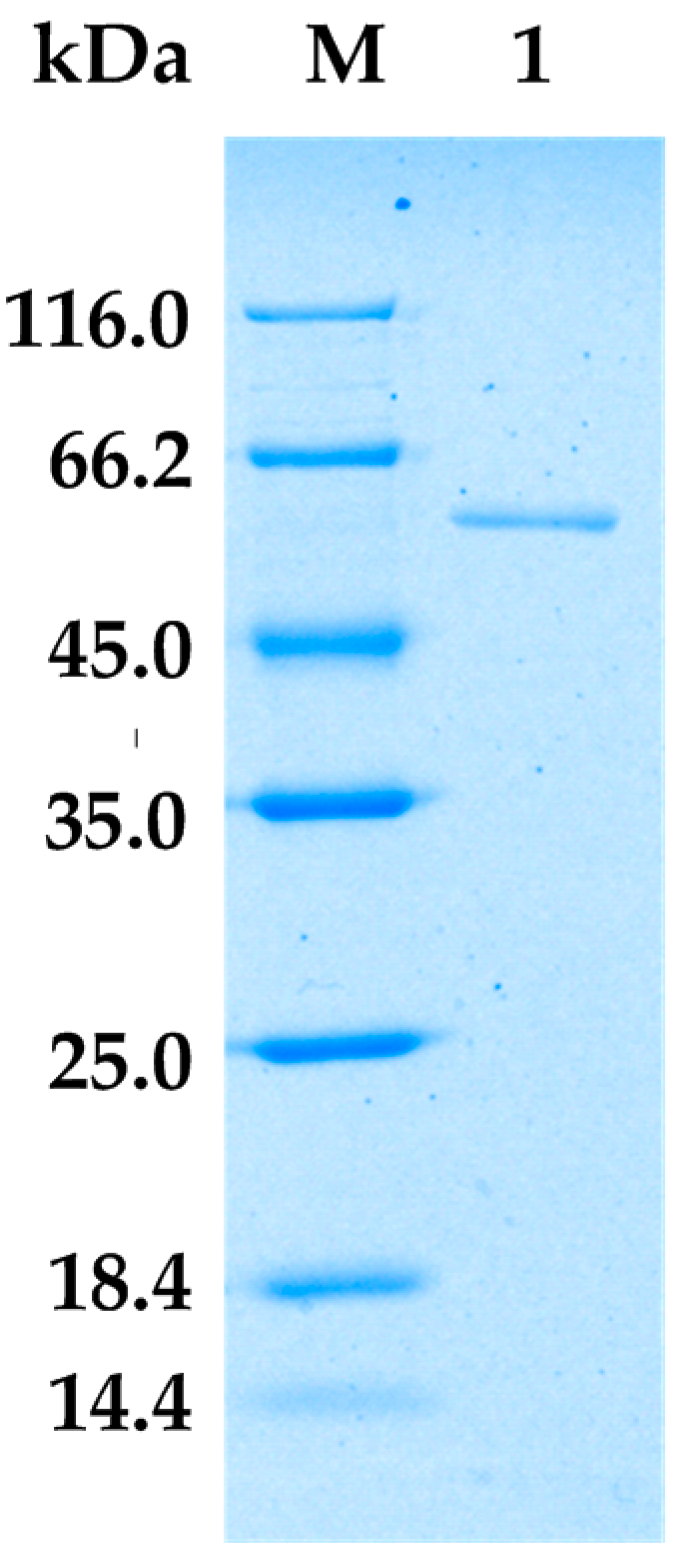
| Step | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Folds | Yield (%) |
|---|---|---|---|---|---|
| Culture broth | 23.82 | 23,724.72 | 996.01 | 1.00 | 100 |
| Vivaflow50 | 10.30 | 22,672.14 | 2201.18 | 2.21 | 95.4 |
| Source 15Q | 1.78 | 14,785.9 | 8306.71 | 8.34 | 62.4 |
| Enzyme | Source | Specific Activity (U/mg) | Molecular Mass (kDa) | Optimal Temperature (°C) | Optimal pH | Cation Activators | Substrate Specificity |
|---|---|---|---|---|---|---|---|
| AlgA | This study | 8306.71 | 60 | 40 | 7.5 | Na+, Mg2+, Ca2+, Mn2+ | PM |
| Oal17 | Shewanella sp.Kz7 [25] | 32 | 82 | 50 | 6.2 | Na+ | PM |
| Cel32 | Cellulophaga sp. NJ-1 [15] | 2417.8 | 32 | 50 | 8.0 | Ca2+, Mg2+, K+ | PM, PG |
| Alg7D | Saccharophagus degradans [26] | 4.6 | 63.2 | 50 | 7.0 | Na+ | PM, PG |
| AlySY08 | Vibrio sp. Aly08 [27] | 1070.2 | 33 | 40 | 7.6 | Na+, K+, Ca2+, Mg2+ | PM, PG |
| AlyV5 | Vibrio sp. QY105 [28] | 2152 | 37 | 38 | 7.0 | Na+, Mg2+, Ca2+, Mn2+ | PM, PG |
| Alm | Agarivorans sp. JAM-Alm [29] | 108.5 | 31 | 30 | 10.0 | Na+, K+ | PG, PMG |
| FlAlyA | Flavabacterium sp. UMI-01 [16] | 2347.8 | 30 | 55 | 7.7 | Na+, K+, Ca2+, Mg2+ | PM |
| Parameter | Sodium Alginate | PolyM |
|---|---|---|
| Vmax (U mg of protein−1) | 1052.0 ± 214.6 | 547.6 ± 22.4 |
| Km (mg mL−1) | 9.0 ± 3.3 | 3.6 ± 0.4 |
| kcat (s−1) | 911.7 ± 185.9 | 474.6 ± 19.4 |
| kcat/Km (mg−1 mL s−1) | 101.3 ± 20.7 | 132.0 ± 5.4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, P.; Zhu, Y.; Men, Y.; Zeng, Y.; Sun, Y. Purification and Characterization of a Novel Alginate Lyase from the Marine Bacterium Bacillus sp. Alg07. Mar. Drugs 2018, 16, 86. https://doi.org/10.3390/md16030086
Chen P, Zhu Y, Men Y, Zeng Y, Sun Y. Purification and Characterization of a Novel Alginate Lyase from the Marine Bacterium Bacillus sp. Alg07. Marine Drugs. 2018; 16(3):86. https://doi.org/10.3390/md16030086
Chicago/Turabian StyleChen, Peng, Yueming Zhu, Yan Men, Yan Zeng, and Yuanxia Sun. 2018. "Purification and Characterization of a Novel Alginate Lyase from the Marine Bacterium Bacillus sp. Alg07" Marine Drugs 16, no. 3: 86. https://doi.org/10.3390/md16030086




