Cytotoxic Polyhydroxysteroidal Glycosides from Starfish Culcita novaeguineae
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Elucidation
2.2. Cytotoxic Activities
3. Experimental Section
3.1. General Experimental Procedures
3.2. Animal Material
3.3. Extraction and Isolation
3.4. Spectral and Physicochemical Data of New Compounds
3.5. Demethylation and Acid Hydrolysis of the New Compounds
3.6. Assays for In Vitro Cytotoxicity
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DEPT | Distortionless enhancement by polarization transfer |
| ESIMS | Electron spray ionization mass spectrum |
| 1H-1H COSY | 1H-1H Correlation spectroscopy |
| HMBC | 1H-detected heteronuclear multiple bond correlation |
| HPLC | High performance liquid chromatography |
| HRESIMS | High resolution electron spray ionization mass spectrum |
| HSQC | 1H-detected heteronuclear single quantum correlation |
| NMR | Nuclear magnetic resonance |
| NOESY | Nuclear overhauser enhancement spectroscopy |
| TLC | Thin layer chromatography |
| TOCSY | Total correlation spectroscopy |
References
- Mah, C.L.; Blake, D.B. Global diversity and phylogeny of the Asteroidea (Echinodermata). PLoS ONE 2012, 7, e35644. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Yi, Y.; Li, L.; Sun, P. Bioactive asterosaponins from the starfish Culcita novaeguineae. J. Nat. Prod. 2005, 68, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Kicha, A.A.; Ivanchina, N.V.; Kalinovsky, A.I.; Dmitrenok, P.S.; Stonik, V.A. Steroidal monoglycosides from the Far Eastern starfish Hippasteria kurilensis and hypothetic pathways of polyhydroxysteroid biosynthesis in starfish. Steroids 2009, 74, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.S.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Fusetani, N. Marine pharmacology in 2009–2011: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of Action. Mar. Drugs 2013, 11, 2510–2573. [Google Scholar] [PubMed]
- Ma, N.; Tang, H.F.; Qiu, F.; Lin, H.W.; Tian, X.R.; Yao, M.N. Polyhydroxysteroidal glycosides from the starfish anthenea chinensis. J. Nat. Prod. 2010, 73, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Ivanchina, N.V.; Kicha, A.A.; Stonik, V.A. Steroid glycosides from marine organisms. Steroids 2011, 76, 425–454. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.X.; Kang, Y.F.; Han, H. Three New Cytotoxic Polyhydroxysteroidal Glycosides from Starfish Craspidaster hesperus. Mar. Drugs 2016, 14, 189. [Google Scholar] [CrossRef] [PubMed]
- Malyarenko, T.V.; Kharchenko, S.D.; Kicha, A.A.; Ivanchina, N.V.; Dmitrenok, P.S.; Chingizova, E.A.; Pislyagin, E.A.; Evtushenko, E.V.; Antokhina, T.I.; Van Minh, C.; et al. Anthenosides L–U, Steroidal Glycosides with Unusual Structural Features from the Starfish Anthenea aspera. J. Nat. Prod. 2016, 79, 3047–3056. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Qi, J.; Ojika, M. Structure–activity relationships of novel neuritogenic steroid glycosides from the Okinawan starfish Linckia laevigata. Bioorg. Med. Chem. 2006, 14, 4458–4465. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Ojika, M.; Sakagami, Y. Linckosides A and B, two new neuritogenic steroid glycosides from the Okinawan starfish Linckia laevigata. Bioorg. Med. Chem. 2002, 10, 1961–1966. [Google Scholar] [CrossRef]
- Kicha, A.A.; Ivanchina, N.V.; Kalinovsky, A.I.; Dmitrenok, P.S.; Sokolova, E.V.; Agafonova, I.G.; Morre, J.; Stonik, V.A. Four new steroid glycosides from the Vietnamese starfish Linckia laevigata. Russ. Chem. Bull. 2007, 56, 823–830. [Google Scholar] [CrossRef]
- Guan, H.S.; Wang, S.G. Chinese Marine Herbal; Shanghai Science and Technology Press: Shanghai, China, 2009; Volume 3, pp. 608–610. [Google Scholar]
- Ngoan, B.T.; Hanh, T.T.H.; Vien, L.T.; Diep, C.N.; Thao, N.P.; Thao, D.T.; Van Thanh, N.; Cuong, N.X.; Nam, N.H.; Thung, D.C.; et al. Asterosaponins and glycosylated polyhydroxysteroids from the starfish Culcita novaeguineae and their cytotoxic activities. J. Asian Nat. Prod. Res. 2015, 17, 1010–1017. [Google Scholar] [CrossRef] [PubMed]
- Iorizzi, M.; Minale, L.; Riccio, R.; Higa, T.; Tanaka, J. Starfish saponins, Part 46.1Steroidal glycosides and polyhydroxysteroids from the starfish Culcita novaeguineae. J. Nat. Prod. 1991, 54, 1254–1264. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.F.; Yi, Y.H.; Li, L.; Sun, P.; Zhang, S.Q.; Zhao, Y.P. Asterosaponins from the starfish Culcita novaeguineae and their bioactivities. Fitoterapia 2006, 77, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.F.; Cheng, G.; Wu, J.; Chen, X.L.; Zhang, S.Y.; Wen, A.D.; Lin, H.W. Cytotoxic asterosaponins capable of promoting polymerization of tubulin from the starfish Culcita novaeguineae. J. Nat. Prod. 2009, 72, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.F.; Yi, Y.H.; Li, L.; Sun, P.; Zhang, S.Q.; Zhao, Y.P. Three New Asterosaponins from the Starfish Culcita novaeguineae and their Bioactivity. Planta Med. 2005, 71, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Zhang, X.; Tang, H.F.; Zhang, Y.; Zhang, X.H.; Cao, W.D.; Gao, D.K.; Wang, X.L.; Jin, B.Q. Asterosaponin 1, a cytostatic compound from the starfish Culcita novaeguineae, functions by inducing apoptosis in human glioblastoma U87MG cells. J. Neurooncol. 2006, 79, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Iorizzi, M.; de Riccardis, F.; Minale, L.; Riccio, R. Starfish saponins, 52. Chemical constituents from the starfish echinaster brasiliensis. J. Nat. Prod. 1993, 56, 2149–2162. [Google Scholar] [CrossRef] [PubMed]
- Kicha, A.A.; Ivanchina, N.V.; Kalinovsky, A.I.; Dmitrenok, P.S.; Smirnov, A.V. Two New Steroid Glycosides from the Far East Starfish Hippasteria kurilensis. Russ. J. Bioorg. Chem. 2009, 35, 504–509. [Google Scholar] [CrossRef]
- Zhu, D.; Yu, B. Total Synthesis of Linckosides A and B, the Representative Starfish Polyhydroxysteroid Glycosides with Neuritogenic Activities. J. Am. Chem. Soc. 2015, 137, 15098–15101. [Google Scholar] [CrossRef] [PubMed]
- Malyarenko, T.V.; Kicha, A.A.; Kalinovsky, A.I.; Ivanchina, N.V.; Popov, R.S.; Pislyagin, E.A.; Menchinskaya, E.S.; Padmakumar, K.P.; Stonik, V.A. Four New Steroidal Glycosides, Protolinckiosides A–D, from the Starfish Protoreaster lincki. Chem. Biodivers. 2016, 998–1007. [Google Scholar] [CrossRef] [PubMed]
- Alley, M.C.; Scudiero, D.A.; Monks, A.; Hursey, M.L.; Czerwinski, M.J.; Fine, D.L.; Abbott, B.J.; Mayo, J.G.; Shoemaker, R.H.; Boyd, M.R. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988, 48, 589–601. [Google Scholar] [PubMed]
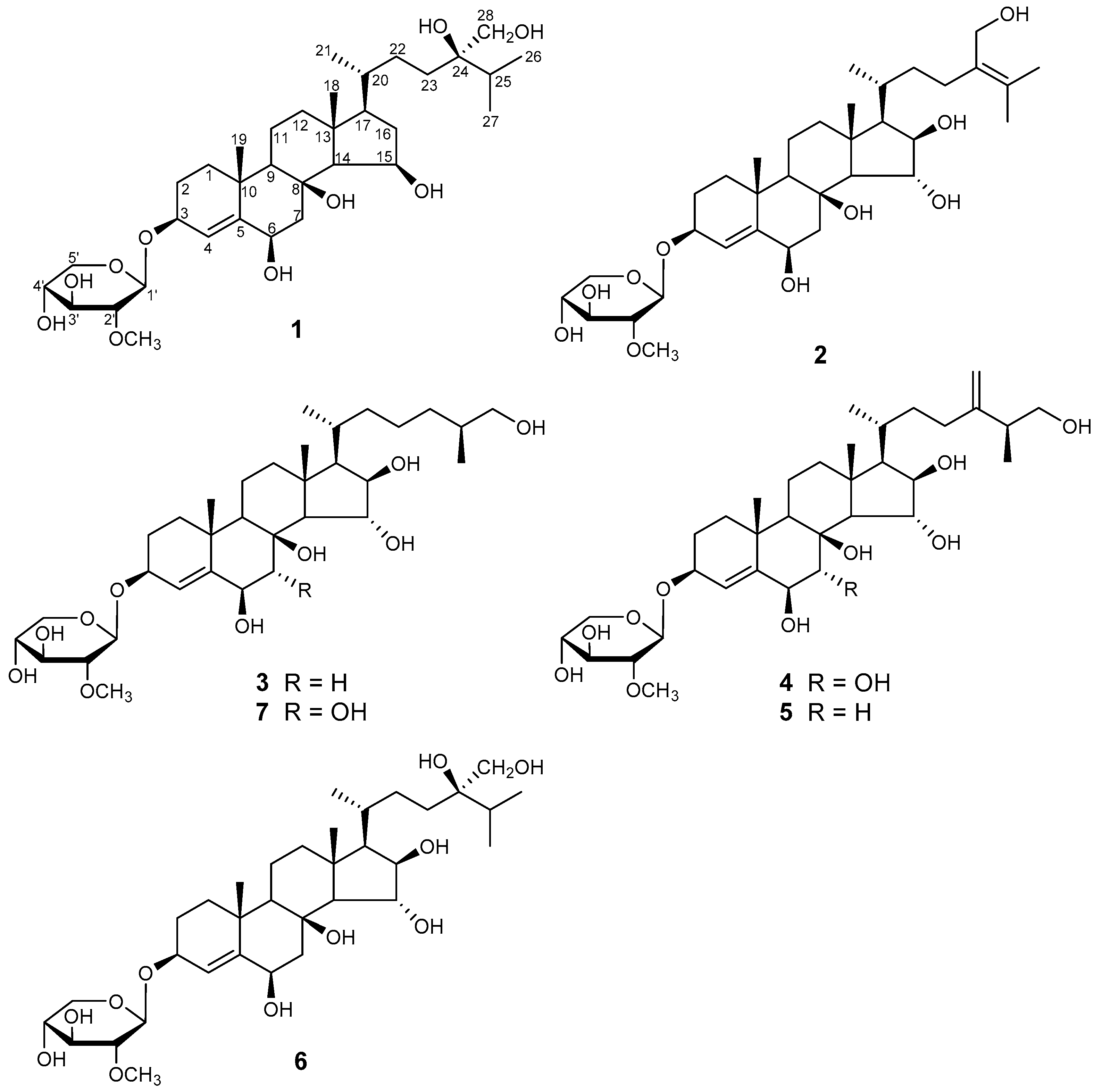
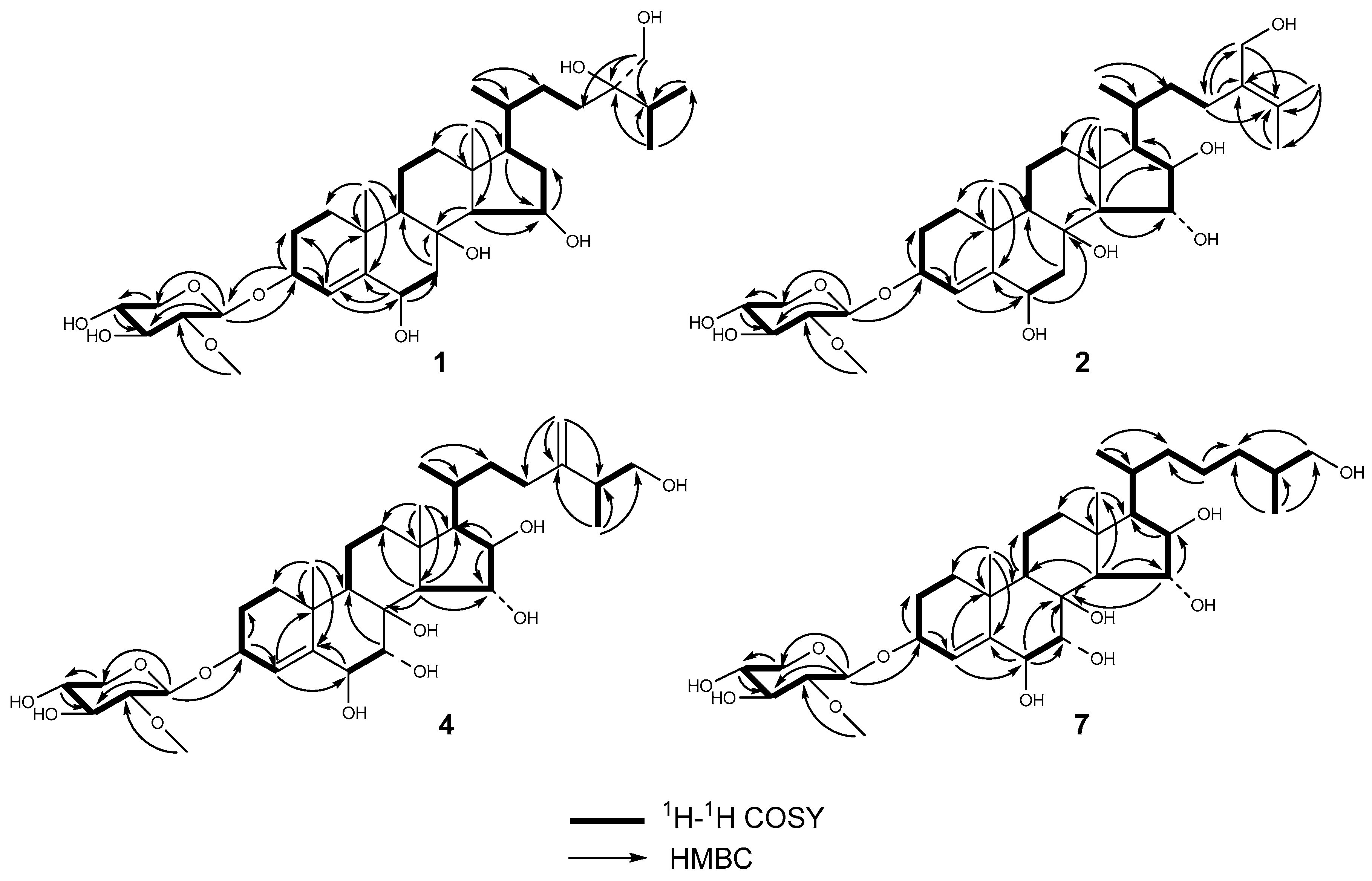
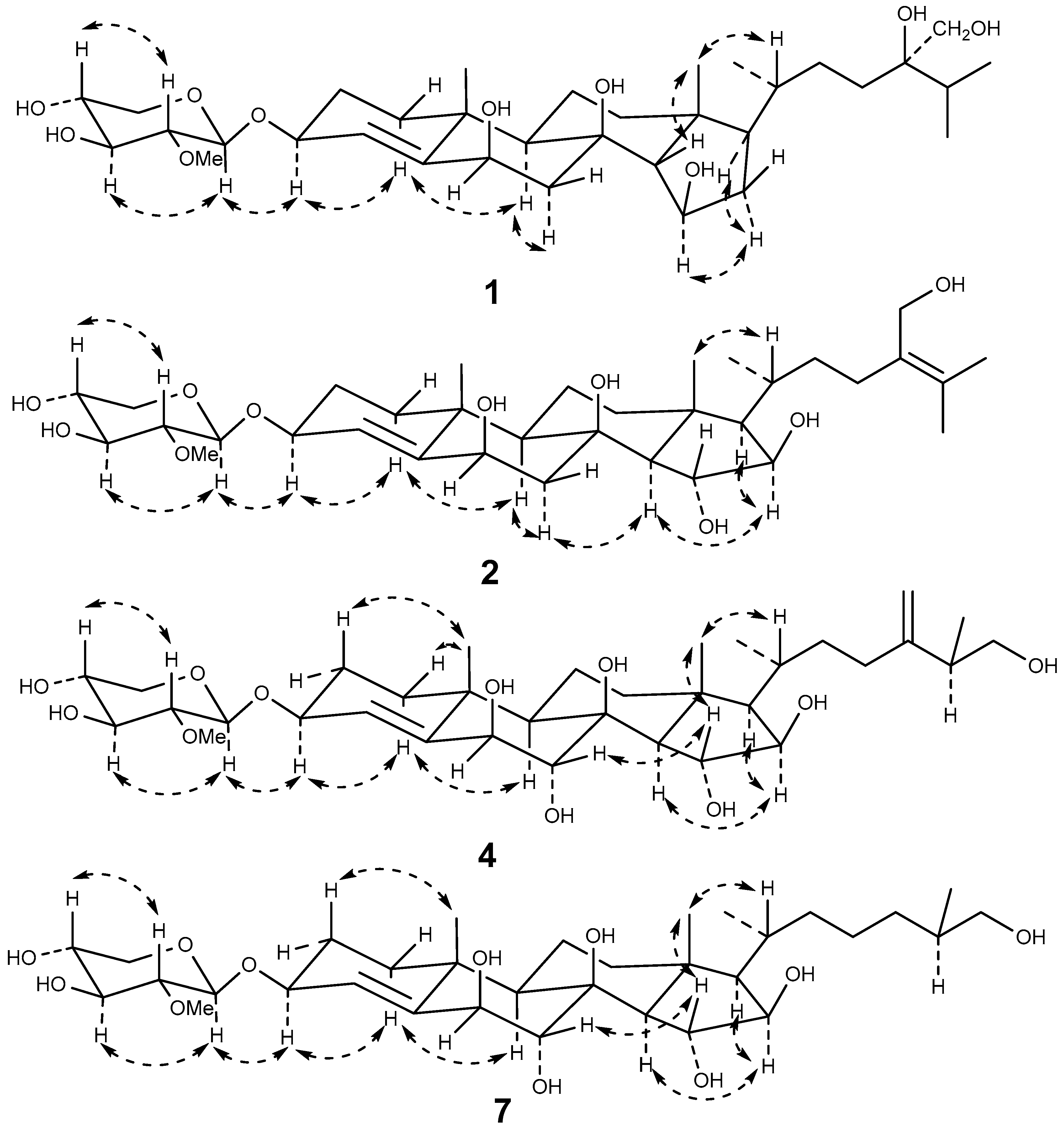
| Position | 1 c,e | 2 d,e | 4 c,f | 7 c,e | ||||
|---|---|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | δC | δH | |
| 1a | 39.4 | 1.85 m | 39.4 | 1.86 m | 39.6 | 1.77 m | 39.4 | 1.90 m |
| 1b | 1.39 m | 1.40 m | 1.32 m | 1.57 m | ||||
| 2a | 28.2 | 2.28 m | 28.2 | 2.28 m | 28 | 1.99 m | 28.3 | 2.28 m |
| 2b | 2.09 m | 2.09 m | 1.76 m | 2.12 m | ||||
| 3 | 76.6 | 4.48 brt (7.2) | 76.7 | 4.48 m | 77.5 | 4.24 brt (8.2) | 76.6 | 4.51 m |
| 4 | 126.5 | 5.94 s | 126.5 | 5.94 s | 130.3 | 5.70 s | 129.8 | 6.17 s |
| 5 | 148.9 | - | 149 | - | 145.4 | - | 146.2 | - |
| 6 | 75.9 | 4.77 d (2.5) | 76 | 4.76 d (2.3) | 79.9 | 4.12 d(3.1) | 80 | 4.87 d(2.7) |
| 7a | 44.9 | 3.40 dd (14.8, 2.5) | 44.9 | 3.49 dd (14.7, 2.3) | 73.9 | 3.96 d(3.1) | 74.5 | 4.84 d(2.7) |
| 7b | 2.07 m | 2.08 m | ||||||
| 8 | 76 | - | 76 | - | 78 | - | 78.1 | - |
| 9 | 57.6 | 1.24 m | 60.8 | 1.55 m | 51.5 | 1.34 m | 51.3 | 1.91 m |
| 10 | 37.6 | - | 37.7 | - | 37.4 | - | 37.3 | - |
| 11a | 19.7 | 2.10 m | 19.6 | 2.15 m | 19.4 | 1.89 m | 19.5 | 2.27 m |
| 11b | 1.55 m | 1.57 m | 1.53 m | 1.67 m | ||||
| 12a | 42.5 | 2.07 m | 43 | 2.14 m | 43.1 | 1.98 m | 43 | 2.16 m |
| 12b | 1.28 m | 1.32 m | 1.22 m | 1.40 m | ||||
| 13 | 45 | - | 45 | - | 45.5 | - | 45.3 | - |
| 14 | 66.6 | 1.56 m | 64.1 | 1.51 m | 59.5 | 1.42 d (10.6) | 59.7 | 2.05 d (10.5) |
| 15 | 69.5 | 4.90 m | 80.8 | 5.08 m | 80.1 | 4.20 dd (10.6, 1.8) | 80.4 | 5.11 brd (10.5) |
| 16a | 42.3 | 2.27 m | 83 | 4.78 m | 82.6 | 4.03 dd (7.4, 1.8) | 82.5 | 4.72 dd (7.1, 1.2) |
| 16b | 2.15 m | |||||||
| 17 | 55.8 | 1.61 brd (9.1) | 60.8 | 1.55 m | 61.4 | 1.29 dd (10.8, 7.4) | 61.6 | 1.54 m |
| 18 | 16 | 1.30 s | 17.5 | 1.74 s | 17 | 1.16 s | 17.6 | 1.77 s |
| 19 | 23.1 | 1.66 s | 23.1 | 1.67 s | 23.2 | 1.34 s | 23.6 | 1.68 s |
| 20 | 36.6 | 1.54 m | 30.6 | 2.42 m | 30.7 | 1.90 m | 30.5 | 2.40 m |
| 21 | 19.4 | 1.07 d (6.2) | 19.1 | 1.21 d (6.7) | 18.5 | 0.96 d (6.7) | 18.8 | 1.14 d (6.7) |
| 22a | 30.6 | 1.92 m | 35.7 | 2.20 m | 35.4 | 1.74 m | 37.1 | 1.96 m |
| 22b | 1.41 m | 1.54 m | 1.23 m | 1.33 m | ||||
| 23a | 32 | 2.03 m | 29.2 | 2.55 t (7.3) | 33 | 2.15 m | 25 | 1.69 m |
| 23b | 1.85 m | 1.52 m | 1.97 m | 1.44 m | ||||
| 24a | 76.2 | - | 135 | - | 154 | - | 35 | 1.64 m |
| 24b | 1.23 m | |||||||
| 25 | 34 | 2.26 m | 128.1 | - | 43.5 | 2.31 m | 37.3 | 1.82 m |
| 26a | 18 | 1.20 d (6.8) | 20.7 | 1.75 s | 67.6 | 3.58 m | 68 | 3.76 dd (10.4, 5.6) |
| 26b | 3.37 m | 3.64 m | ||||||
| 27 | 18.1 | 1.22 d (6.8) | 21.1 | 1.77 s | 17.4 | 1.07 d (6.5) | 18 | 1.08 d (6.7) |
| 28a | 66.1 | 4.03 m | 62.4 | 4.51 m | 109.4 | 4.84 s | - | - |
| 28b | 3.97 m | 4.42 m | 4.77 s | |||||
| 2-OMe-Xyl | ||||||||
| 1′ | 104.8 | 4.84 d (7.6) | 104.8 | 4.84 d (7.6) | 104.7 | 4.44 d (7.6) | 104.7 | 4.76 d (7.6) |
| 2′ | 85.6 | 3.46 dd (8.8, 7.6) | 85.6 | 3.46 dd (8.8, 7.6) | 85 | 2.85 dd (9.0, 7.6) | 85.5 | 3.44 dd (8.9, 7.6) |
| 3′ | 78.2 | 4.08 m | 78.2 | 4.08 br.t(8.8) | 77.6 | 3.33 m | 78.1 | 4.04 br.t (8.9) |
| 4′ | 71.6 | 4.23 m | 71.6 | 4.23 m | 71.4 | 3.50 m | 71.6 | 4.20 m |
| 5′a | 67.5 | 4.35 dd (11.3, 5.4) 3.67 m | 67.5 | 4.34 dd (11.4, 5.4) | 66.9 | 3.84 dd (11.5, 5.4) | 67.5 | 4.32 dd (11.3, 5.3) |
| 5′b | 3.67 t (11.4) | 3.18dd (11.5, 10.4) | 3.62 m | |||||
| 2-OMe | 61.2 | 3.72 s | 61.2 | 3.72 s | 61.3 | 3.59 s | 61.1 | 3.68 s |
| Compounds | Cytotoxic Activity (IC50, µM) | ||
|---|---|---|---|
| U87 | U251 | SHG44 | |
| 1 | 9.35 ± 0.46 | 11.28 ± 0.65 | 8.04 ± 0.32 |
| 2 | 33.52 ± 1.23 | 40.76 ± 1.58 | 36.54 ± 1.44 |
| 4 | 26.33 ± 1.16 | 22.66 ± 1.28 | 35.26 ± 1.51 |
| 7 | 43.25 ± 1.73 | 28.93 ± 1.83 | 26.22 ± 1.64 |
| Doxorubicin | 0.33 ± 0.02 | 0.24 ± 0.01 | 0.15 ± 0.01 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Li, H.; Wang, M.; Liu, Y.; Feng, Y.; Liu, K.; Tang, H. Cytotoxic Polyhydroxysteroidal Glycosides from Starfish Culcita novaeguineae. Mar. Drugs 2018, 16, 92. https://doi.org/10.3390/md16030092
Lu Y, Li H, Wang M, Liu Y, Feng Y, Liu K, Tang H. Cytotoxic Polyhydroxysteroidal Glycosides from Starfish Culcita novaeguineae. Marine Drugs. 2018; 16(3):92. https://doi.org/10.3390/md16030092
Chicago/Turabian StyleLu, Yunyang, Hu Li, Minchang Wang, Yang Liu, Yingda Feng, Ke Liu, and Haifeng Tang. 2018. "Cytotoxic Polyhydroxysteroidal Glycosides from Starfish Culcita novaeguineae" Marine Drugs 16, no. 3: 92. https://doi.org/10.3390/md16030092




