Advances in Marine Microbial Symbionts in the China Sea and Related Pharmaceutical Metabolites
Abstract
:1. Introduction
2. Diversity revelation of marine microbial symbionts in the China Sea
3. Biological activity and pharmaceutical metabolites of marine microbial symbionts in the China Sea
4. Screening of functional gene from marine microbial symbionts in the China Sea
5. Concluding remarks and perspectives
Acknowledgements
References and Notes
- http://en.wikipedia.org/wiki/Symbiosis.
- Zhang, H; Lee, YK; Zhang, W; Lee, HK. Culturable actinobacteria from the marine sponge Hymeniacidon perleve: isolation and phylogenetic diversity by 16S rRNA gene-RFLP analysis. Antonie van Leeuwenhoek 2006, 90, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y; Huang, J; Deng, M; Zhang, W. Culture-independent nested PCR method reveals high diversity of actinobacteria associated with the marine sponges Hymeniacidon perleve and Sponge sp. Antonie van Leeuwenhoek 2008, 94, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H; Zheng, W; Huang, J; Luo, H; Jin, Y; Zhang, W; Liu, Z; Huang, Y. Actinoalloteichus hymeniacidonis sp. nov., an actinomycete isolated from the marine sponge Hymeniacidon perleve. International Journal of Systematic and Evolutionary Microbiology 2006, 56, 2309–2312. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S; Li, X; Zhang, L; Sun, W; Dai, S; Xie, L; Liu, Y; Lee, KJ. Culturable actinobacteria isolated from marine sponge Iotrochota sp. Marine Biology 2008, 153, 945–952. [Google Scholar] [CrossRef]
- Jiang, S; Sun, W; Chen, M; Dai, S; Zhang, L; Liu, Y; Lee, KJ; Li, X. Diversity of culturable actinobacteria isolated from marine sponge Haliclona sp. Antonie van Leeuwenhoek 2007, 92, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Li, Z; He, L; Wu, J; Jiang, Q. Bacterial community diversity associated with four marine sponges from the South China Sea based on 16S rDNA-DGGE fingerprinting. Journal of Experimental Marine Biology and Ecology 2006, 329, 75–85. [Google Scholar] [CrossRef]
- Li, Z; Liu, Y. Marine sponge Craniella austrialiensis-associated bacterial diversity revelation based on 16S rDNA library and biologically active actinomycetes screening, phylogenetic analysis. Letters in Applied Microbiology 2006, 43, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H; Zhang, W; Jin, Y; Jin, M; Yu, X. A comparative study on the phylogenetic diversity of culturable actinobacteria isolated from five marine sponge species. Antonie van Leeuwenhoek 2008, 93, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z; Zeng, W; Huang, Y; Yang, Z; Li, J; Cai, H; Su, W. Detection of antitumor and antimicrobial activities in marine organism associated actinomycetes isolated from the Taiwan Strait, China. FEMS Microbiology Letters 2000, 188, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Li, Z; Hu, Y; Liu, Y; Huang, Y; He, L; Miao, X. 16S rDNA clone library based bacterial phylogenetic diversity associated with three South China Sea sponges. World Journal of Microbiology and Biotechnology 2007, 23, 1265–1272. [Google Scholar] [CrossRef]
- Li, Z; Hu, Y; Huang, Y; Huang, Y. Isolation and phylogenetic analysis of the biologically active bacteria associated with three south China sea sponges. Microbiology 2007, 76, 494–499. [Google Scholar] [CrossRef] [PubMed]
- Li, Z; He, L; Miao, X. Cultivable bacterial community from South China Sea sponge as revealed by DGGE fingerprinting and 16S rDNA phylogenetic analysis. Current Microbiology 2007, 55, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Lee, OO; Lau, SCK; Qian, P. Consistent bacterial community structure associated with the surface of the sponge. Mycale adhaerens Bowerbank Microbial Ecology 2006, 52, 693–707. [Google Scholar] [CrossRef]
- Qian, PY; Dobretsov, S; Dahms, HU; Pawlik, J. Antifouling activity and microbial diversity of two congeneric sponges Callyspongia spp. from Hong Kong and the Bahamas. Marine Ecology Progress Series 2006, 324, 151–165. [Google Scholar] [CrossRef]
- Zheng, L; Han, X; Chen, H; Yan, XJ. Marine bacteria associated with marine macroorganisms: the potential antimicrobial resources. Annals of Microbiology 2005, 55, 119–124. [Google Scholar]
- Dobretsov, S; Qian, P. The role of epibotic bacteria from the surface of the soft coral Dendronephthya sp. in the inhibition of larval settlement. Journal of Experimental Marine Biology and Ecology 2004, 299, 35–50. [Google Scholar] [CrossRef]
- Harder, T; Lau, CKS; Dobretsov, S; Fang, Tsz K; Qian, P-Y. A distinctive epibiotic bacterial community on the soft coral Dendronephthya sp. and antibacterial activity of coral tissue extracts suggest chemical mechanism against bacterial epibiosis. FEMS Microbiology Ecology 2003, 43, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N; Fang, Z; Huang, H; Fang, Z; Wu, X; Bao, S. Phylogenetic diversity of bacteria and archeaea associated with the marine sponge Pachychalina sp. Polish Journal of Ecology 2008, 56, 505–510. [Google Scholar]
- Zheng, L; Chen, H; Han, X; Lin, W; Yan, X. Antimicrobial screening and active compound isolation from marine bacterium NJ6-3-1 associated with the sponge. Hymeniacidon perleve World Journal of Microbiology & Biotechnology 2005, 21, 201–206. [Google Scholar] [CrossRef]
- Li, Z; Peng, C; Shen, Y; Miao, X; Zhang, H; Lin, H. L,L-diketopiperazines from Alcaligens feacalis A72 associated with South China Sea sponge. Stelletta tenuis Biochemical Systematics and Ecology 2008, 36, 230–234. [Google Scholar] [CrossRef]
- Liu, R; Cui, C; Duan, L; Gu, Q; Zhu, W. Potent in vitro anticancer activity of metacycloprodigiosin and undecylprodigiosin from a sponge-derived actinomycete Saccharopolyspora sp. nov. Archives of Pharmacal Research 2005, 28, 1341–1344. [Google Scholar] [CrossRef] [PubMed]
- Jeong, SY; Ishida, K; Ito, Y; Okada, S; Murakami, M. Bacillamide, a novel algicide from the marine bacterium, Bacillus sp. SY-1, against the harmful dinoflagellate, Cochlodinium polykrikoides. Tetrahedron Letters 2003, 44, 8005–8007. [Google Scholar] [CrossRef]
- Yu, L; Li, Z; Peng, C; Li, Z; Guo, Y. Neobacillamide A, a novel thiazole-containing alkaloid from the marine bacterium, Bacillus vaiismortis C89, that was associated with South China Sea sponge Dysidea avara. Helvetica Chimica Acta 2009. In press. [Google Scholar]
- Xin, ZH; Zhu, WM; Gu, QQ; Fang, YC; Duan, L; Cui, CB. A new cytotoxic compound from Penicillium auratiogriseum, symbiotic or epiphytic fungus of sponge Mycale plumose. Chinese Chemical Letters 2005, 16, 1227–1229. [Google Scholar]
- Xie, LW; Jiang, SM; Zhu, HH; Sun, W; Ouyang, YC; Dai, SK; Li, X. Potential inhibitors against Sclerotinia sclerotiorum, produced by the fungus Myrothecium sp. associated with the marine sponge Axinella sp. European Journal of Plant Pathology 2008, 122, 571–578. [Google Scholar] [CrossRef]
- Yang, LH; Miao, L; Lee, OO; Li, X; Xiong, H; Pang, K; Virjmoed, L; Qian, P. Effect of culture conditions on antifouling compound production of a sponge-associated fungus. Applied Microbiology Biotechnology 2007, 74 , 1221–1231. [Google Scholar]
- Xin, Z; Fang, Y; Du, L; Zhu, T; Duan, L; Chen, J; Gu, Q; Zhu, W. Aurantiomides A–C, quinazoline alkaloids from the sponge-derived fungus Penicillium aurantiogriseum SP0-19. Journal of Natural Products 2007, 70, 853–855. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z; Tian, L; Zhu, T; Wang, W; Du, L; Fang, Y; Gu, Q; Zhu, W. Isocoumarin derivatives from the sea squirt-derived fungus Penicillium stoloniferum QY2-10 and the halotolerant fungus Penicillium notatum B-52. Archives of Pharmacal Research 2007, 30, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Wang, F; Fang, Y; Zhang, M; Lin, A; Zhu, T; Gu, Q; Zhu, W. Six new ergosterols from the marine-derived fungus Rhizopus sp. Steroids 2008, 73, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Wang, F; Fang, Y; Zhu, T; Zhang, M; Lin, A; Gu, Q; Zhu, W. Seven new prenylated indole diketopiperazine alkaloids from holothurianderived fungus Aspergillus fumigatus. Tetrahedron 2008, 64, 7986–7991. [Google Scholar] [CrossRef]
- Yang, R; Li, C; Lin, Y; Peng, G; She, Z; Zhou, S. Lactones from a brown alga endophytic fungus (No. ZZF36) from the South China Sea and their antimicrobial activities. Bioorganic & Medicinal Chemistry Letters 2006, 16, 4205–4208. [Google Scholar] [CrossRef]
- Zhang, Y; Li, XM; Wang, CY; Wang, BG. A new naphthoquinoneimine derivative from the marine algal-derived endophytic fungus Aspergillus niger EN-13. Chinese Chemical Letters 2007, 18, 951–953. [Google Scholar] [CrossRef]
- Zhang, Y; Wang, S; Li, X; Cui, C; Feng, C; Wang, B. New sphingolipids with a previously unreported 9-methyl-C20 sphingosine moiety from a marine algous endophytic fungus Aspergillus niger EN-13. Lipids 2007, 42, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y; Li, X; Wang, B. Nigerasperones A-C, new monomeric and dimeric naphtha-γ-pyrones from a marine alga-derived endophytic fungus Aspergillus niger EN-13. Journal of Antibiotics 2007, 60, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Wang, S; Li, X; Teuscher, F; Li, D; Diesel, A; Ebel, R; Proksch, P; Wang, B. Chaetopyranin, a benzaldehyde derivative, and other related metabolites from Chaetomium globosum, an endophytic fungus derived from the marine red alga Polysiphonia urceolata. Journal Natural Products 2006, 69, 1622–1625. [Google Scholar] [CrossRef]
- Han, Y; Yang, B; Zhang, F; Miao, X; Li, Z. Characterization of antifungal chitinase from marine Streptomyces sp. DA11 associated with South China Sea sponge Craniella australiensis. Marine Biotechnology 2009, 11, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Han, Y; Li, Z; Miao, X; Zhang, F. Statistical optimization of medium components to improve the chitinase activity of Streptomyces sp. DA11 associated with the South China Sea sponge Craniella australiensis. Process Biochemistry 2008, 43, 1088–1093. [Google Scholar] [CrossRef]
- Zhang, W; Li, Z; Miao, X; Zhang, F. The screening of antimicrobial bacteria with diverse novel nonribosomal peptide synthetase (NRPS) genes from South China Sea sponges. Marine Biotechnology 2009, 11, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W; Zhang, F; Li, Z; Miao, X; Meng, Q; Zhang, X. Investigation of sponge-associated cultivable bacteria with polyketide synthase genes and antimicrobial activity in the South China Sea. Journal of Applied Microbiology 2009. [CrossRef]
| Host | Microorganism | Ref. |
|---|---|---|
| Actinomycetes | ||
| Sponge Hymeniacidon perleve | Actinoalloteichus cyanogriseus, Actinoalloteichus hymeniacidonis sp. nov., Acidimicrobium, Actinomyces, Corynebacterium, Micromonospora aurantiaca, Micrococcus sp., microbacterium, Nocardiopsis dassonvillei, Nocardiopsis lucentensis, Nocardia salmonicida, Rhodococcus opacus, Propionibacterium, Pseudonocardia antarctica, Sporichthya sp., Streptomyces argenteolus, Streptomyces aureofaciens, Streptomyces caviscabies, Streptomyces coelicolor, Streptomyces gibsonii, Streptomyces gougerotii, Streptomyces paradoxus, Streptomyces rimosus, Streptomyces tendae | [2,3,4] |
| Sponge lotrochota sp. | Cellulosimicrobium cellulans, Nocardiopsis, Streptomyces bikiniensis, Streptomyces fradiae, Streptomyces geysiriensis, Streptomyces griseoflavus, Streptomyces griseus subsp., Streptomyces intermedius, Streptomyces sindenensis, Streptomyces parvus, Streptomyces variabilis | [5] |
| Sponge Halidona sp. | Micromonospora carbonacea, Micromonospora floridensis, Micromonospora sp., Nocardiopsis sp., Streptomyces fradiae, Streptomyces griseoincarnatus, Streptomyces variabilis, Verrucosispora gifhornensis | [6] |
| Sponge Craniella australiensis | Actinomycetales bacterium, Streptomyces sp., Pseudonocardia sp., uncultured actinobacterium | [7, 8, 9] |
| Sponges Stelletta tenuis, Halichondria rugosa, Reniochalina sp. | Streptomyces sp., Pseudonocardia sp. | [9] |
| Sponge Sponge sp. | Acidimicrobium, Cellulosimicrobium, Mycobacterium, Nocardia sp., Streptomyces sp., Pseudonocardia sp. | [3,9] |
| Sea hare Aplysia dactylomela, marine alga Gracilaria verrucosa | Micromonospora sp., Streptomyces sp. | [10] |
| Bacteria | ||
| Sponge Stelletta tenuis | Adelie penguin guano bacterium, Alcaligenes faecalis, Acinetobacter johnsonii, Bacillus cereus, Bacillus firmus, Bacillus sporothermodurans, Bdellovibrio sp., Brevundimonas vesicularis, Halomonas sp., Idiomarina sp., Marinomonas sp., Oceanisphaera litoralis, Oleiphilus messinensis, Psychrobacter glacincola, Psychrobacter maritimus, Psychrobacter psychrophilus, Psychrobacter luti, Shewanella pacifica, Sporosarcina sp., Staphylococcus epidermidis, Staphylococcus lentus, Vitellibacter vladivostokensis, uncultured Arctic sea ice bacterium, uncultured Antarctic sea ice bacterium, uncultured alpha-Protebacterium, uncultured bacterium, uncultured Clostridia bacterium | [7, 11, 12, 13] |
| Sponge Halichondria rugosa | Acinetobacter johnsonii, Acidovorax sp., Alcaligenes sp., Bacillus anthracis, Bacillus licheniformis, Bacillus psychrophilus, Bacillus subtilis, Brevundimonas vesicularis, Halomonas sp., Idiomarina sp. NT, Marinomonas sp., Moellerella wisconsensis, Psychrobacter maritimus, Psychrobacter psychrophilus, Pseudomonas sp., Paracoccus halodenitrificans, Providencia sp., Rhodobacteraceae bacterium, Stenotrophomonas maltophilia, marine bacterium KMM 3937, rainbow trout intestinal bacterium, uncultured gamma-Proteobacterium, uncultured alpha-Proteobacterium, uncultured bacterium | [7, 11, 12, 13] |
| Sponge Dysidea avara | Acidovorax sp., Alcaligenes sp., Arenibacter latericius, Bellia baltica, Bacillus cereus, Biziolla paragoriae, Bizionia paragorgiae, Bacillus vallismortis, Brevundimonas vesicularis, Erythobacter lutedus, Flavobacteriaceae str., Halomonas sp., Idiomarina sp., Klebsiella pneumoniae, Marinobacter sp., Oceanisphaera koreensis, Oceanisphaera sp., Psychrobacter cibarius, Psychrobacter fozii, Psychrobacter cibarius, Psychrobacter psychrophilus, Psychrobacter sp., Paracoccus sp., Rhizobiaceae bacterium, Staphylococcus epidermidis, uncultured sponge symbiont, uncultured bacteroidetes bacterium, uncultured Arctic sea ice bacterium, uncultured Pseudoalteromonas sp., uncultured proteobacterium | [7, 11, 12, 13] |
| Sponge Craniella australiensis | Aequorivitaferruginea, Alcaligenes sp., Bacillus sp., Bizionia paragorgiae, Brevundimonas vesicularis, Carnobacterium funditum, Cytophaga sp., Enterobacter hormaechei subsp., Halomonas sp., Hyphomicrobium sp., Ochrobactrum anthropi, Rhodobacter sp., Photorhabdus luminescens, Planococcus sp., Pseudoalteromonas sp., Psychrobacter glacincola, Psychrobacter sp. Roseavarius crassostreae, Staphylococcus sp., Steigerwaltii sp., uncultured Arctic sea ice bacterium, uncultured soil bacterium, uncultured beta-proteobacterium, uncultured gram-positive bacterium | [7,8,13] |
| Sponge Mycale adhaerens | Alteromonas alvinellae, Kocuria rhizophila, Mesophilum sp., Microbulbifer hydrolyticus, Micrococcus kristinae, Pseudoalteromonas piscicida, Pseudoalteromonas spongiae, Shewanella alga, Staphylococcus cohnii, Tenacibaculum sp., Vibrio fluvialis, Vibrio furnissii, Vibrio halioticoli, Vibrio nereis | [14] |
| Sponge Callyspongia sp. | Alteromonas macleodii, Alteromonas marina, Bacillus cereus, Bacillus hwajinpoensis, Bacillus megaterium, Bacillus thuringiensis serovar konkukian, Erythrobacter flavus, Exiguobacterium gaetbuli, Exiguobacterium marinum, Marinobacter aquaeolei, Micrococcus luteus, Planococcus citreus, Pseudovibrio denitrificans, Psychrobacter sp., Ruegeria atlantica, Silicibacter lacuscaerulensis, Vibrio aestuarianus, Vibrio corallilyticus, Vibrio fischeri, Vibrio harveyi, Vibrio hollisae, Vibrio natriegens, Vibrio tubiashi, Vibrio probioticus | [15] |
| Sponge Hymeniacidon perleve Coral Eupexaura curvata Seaweeds Gymnogongrus flabelliformis, Laurencia flabelliformis, Laminaria japonica, Lomentaria catenata, Sargassum thunbergii, Ulvafasciata, Ulvapertusa | Alteromonas sp, Bacillus sp., Flavobacterium sp., Pseudomonas sp. | [16] |
| Soft coral Dendronephthya sp. | Flexibacter sp., Pseudoalteromonas sp., Tenacibaculum mesophilum, Vibrio ichthyoenteri | [17,18] |
| Archaea | ||
| Sponge Pachychalina sp. | Archaeal clone | [19] |
| Sponge Theonella swinhoei | Cenarchaeum | Data to be published |
| Fungi | ||
| Sponge Phakellia fusca | Ascomycota sp., Aspergillns candidus, Aspergillus fiimigatus, Aspergillus ochraceus, Apiospora montagnei, Candi da parapsilosis, Cladosporium sp., Davidiella tassiana, Didymocrea sa dasivanii, Fusarium sp., Hypo crea koningii, Lentomitella cir rhosa, Marasmius alii aceus, Nigr ospora or yzae, P aecilomyces li lacinus, Penicillium chrysogenum, Penicillium purpurogenum, Pestalotiopsis guepinii, Scopu lariopsis brevicaulis, Rhizomucor pusillus | Data to be published |
| Sponge Theonella swinhoei | Ascomycota sp., Aspergillus versicolor, Davidiella tassiana, Fusariu m sp., Paecilomyces lilacinus, Penicillium chrysogenum, Penicillium pinophilum | Data to be published |
| Host | Microorganism | Compound | Bioactivity | Ref. |
|---|---|---|---|---|
| Sponge Hymeniacidon perleve | Pseudoalteromonas piscicida |  | Antimicrobial activity | [20] |
| Sponge Stelletta tenuis | Alcaligenes faecalis A72 | 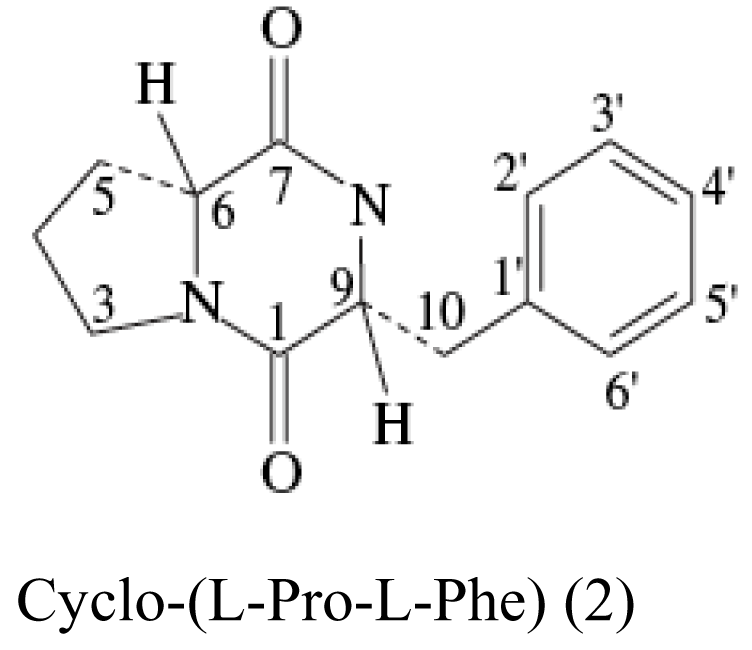 | Moderate antimicrobial activity | [21] |
| Sponge Mycale plumose | Saccharopolyspora sp. nov. | 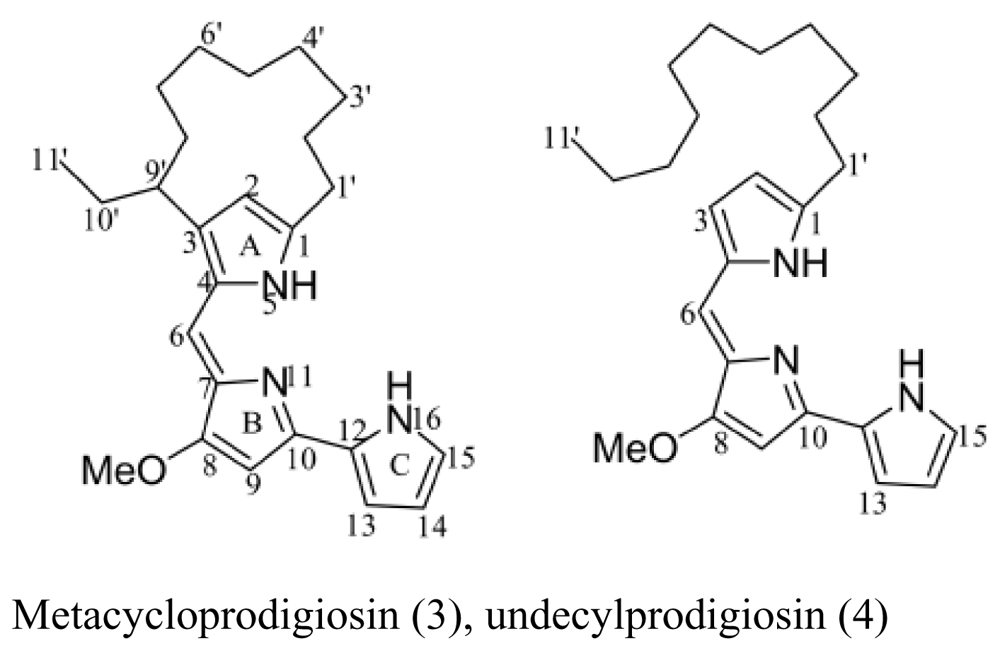 | Cytotoxic activity | [22] |
| Sponge Dysidea avara | Bacillus vallismortis C89 |  | Red-tide algae inhibited | [23,24] |
| Sponge Mycale plumose | Penicillium auratiogriseum | 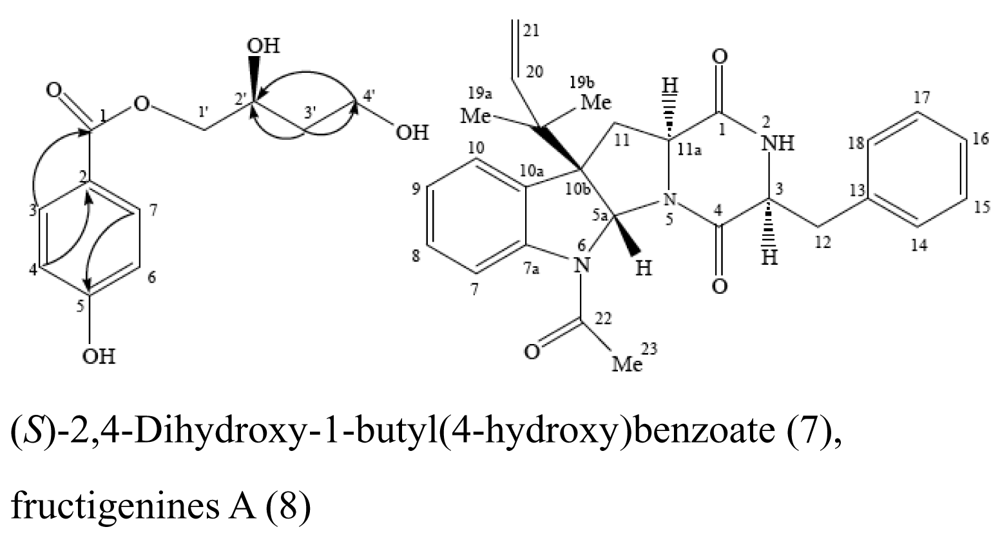 | Cytotoxic activity | [25] |
| Sponge Axinella sp. | Myrothecium sp. | 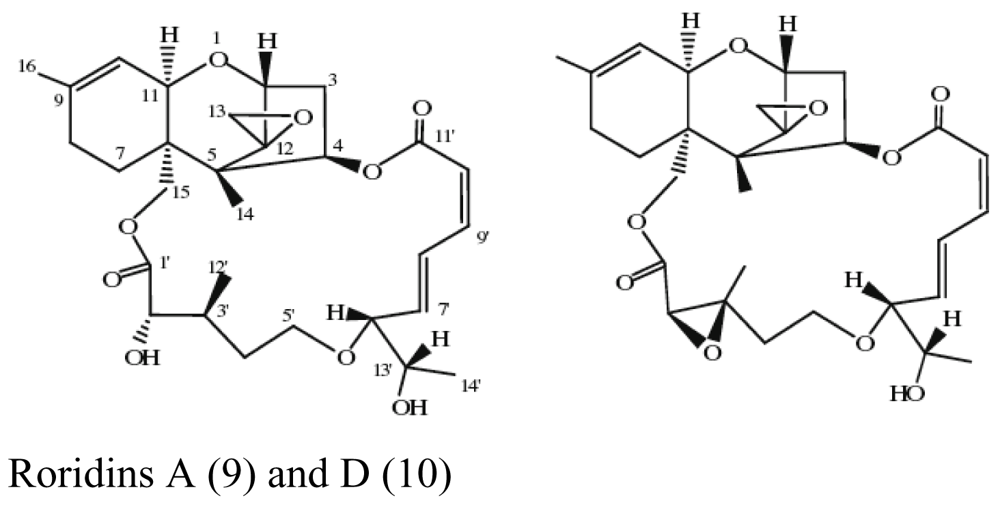 | Antifungal activity | [26] |
| Sponge Acanthella cavernosa | Letendraea helminthicola | 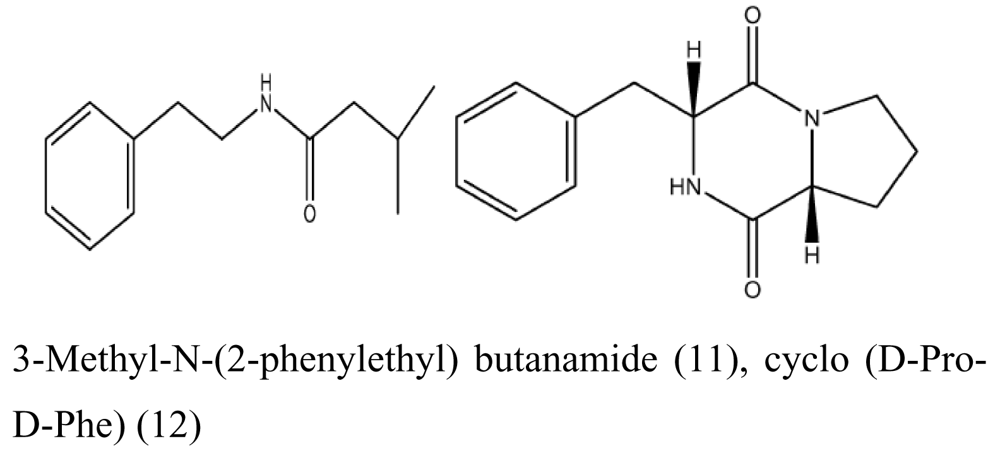 | Antifouling | [27] |
| Sponge Mycale plumose | Penicillium aurantiogriseum SP0-19 | 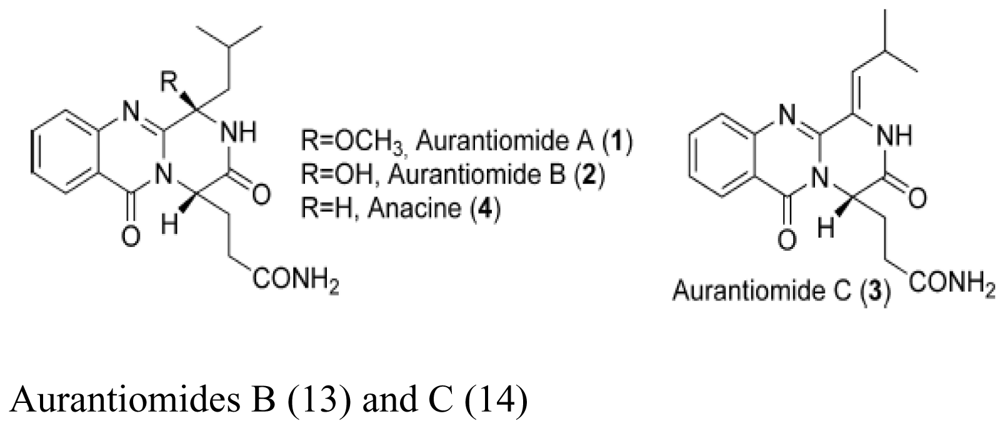 | Moderate cytotoxic activity | [28] |
| Sea squirt (unidentified) | Penicillium stoloniferum QY2-10 | 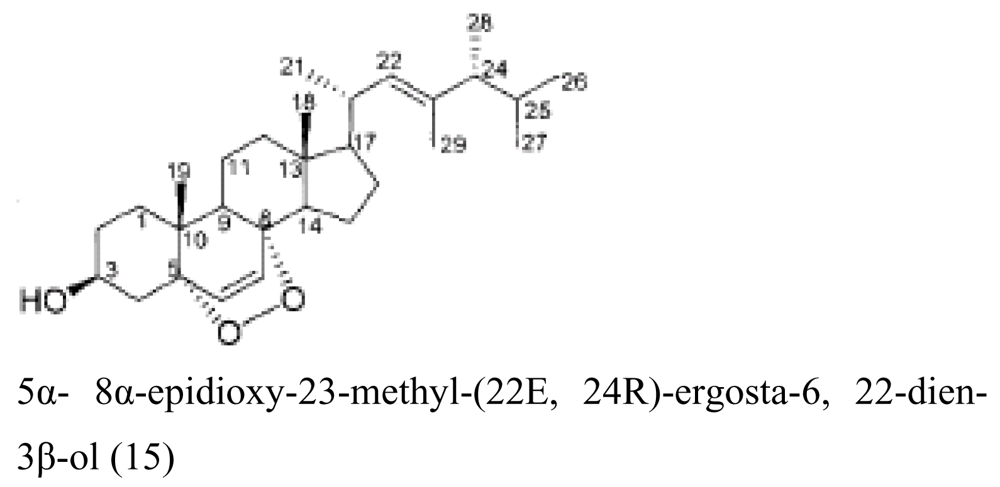 | Cytotoxic activity | [29] |
| Marine Bugula sp. | Rhizopus sp. |  | Cytotoxic activity | [30] |
| Holothurian | Aspergillus fumigatus |  | Cytotoxic activity | [31] |
| Brown alga Sargassum sp. | Unidentified endophytic fungus No. ZZF36 | 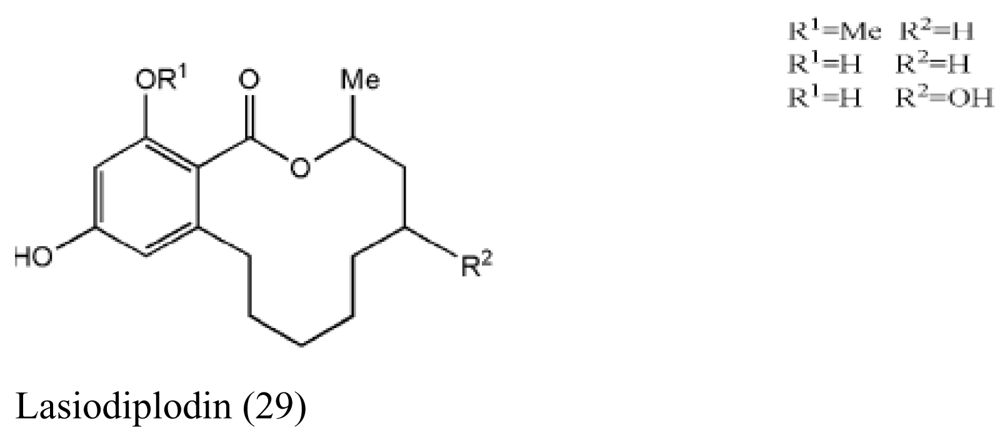 | Antimicrobial activity | [32] |
| Brown alga Colpomenia sinuosa | Aspergillus niger EN-13 | 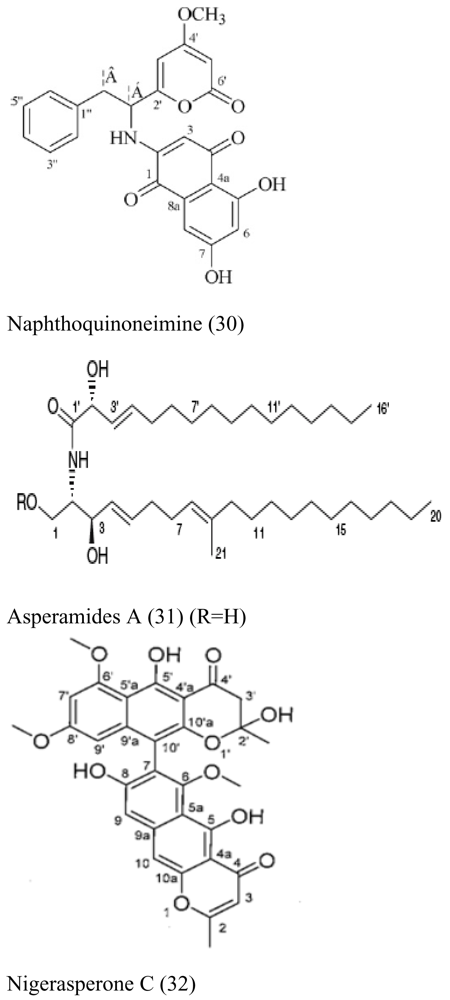 | Moderate antifungal activity | [33,34,35] |
| Red alga Polysiphonia urceolata | Chaetomium globosum | 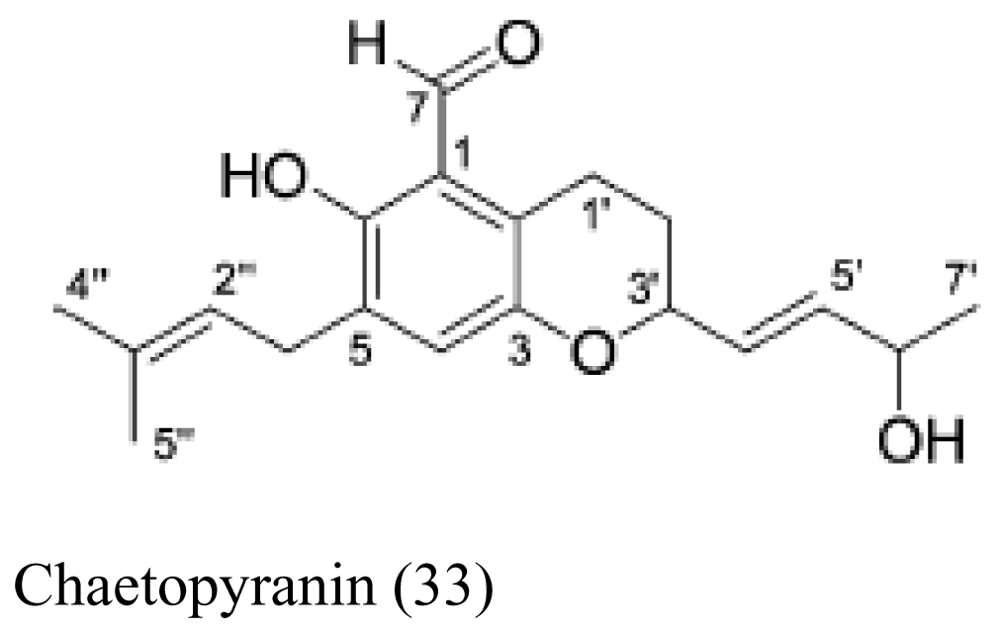 | Moderate to weak cytotoxic activity | [36] |
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, Z. Advances in Marine Microbial Symbionts in the China Sea and Related Pharmaceutical Metabolites. Mar. Drugs 2009, 7, 113-129. https://doi.org/10.3390/md7020113
Li Z. Advances in Marine Microbial Symbionts in the China Sea and Related Pharmaceutical Metabolites. Marine Drugs. 2009; 7(2):113-129. https://doi.org/10.3390/md7020113
Chicago/Turabian StyleLi, Zhiyong. 2009. "Advances in Marine Microbial Symbionts in the China Sea and Related Pharmaceutical Metabolites" Marine Drugs 7, no. 2: 113-129. https://doi.org/10.3390/md7020113




