Deoxyamphimedine, a Pyridoacridine Alkaloid, Damages DNA via the Production of Reactive Oxygen Species
Abstract
:1. Introduction
2. Results and Discussion
2.1. Cytotoxicity in Cultured Mammalian Cell Lines
2.2. Deoxyamphimedine Intercalates DNA
2.3. Deoxyamphimedine Cleaves DNA Independently of Topoisomerase 1 or 2
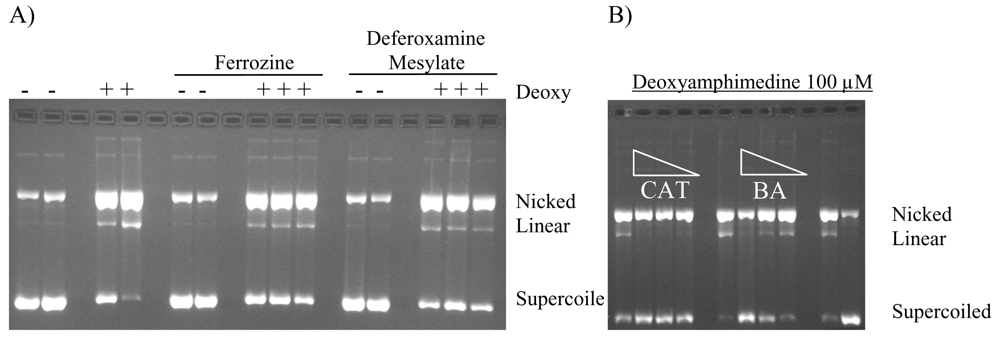
2.4. Protection of DNA from Deoxyamphimedine Cleavage
2.5. Discussion
3. Experimental
3.1. Chemicals and Reagents
3.2. Assessment of DNA Intercalation
3.3. Quantitation of DNA and Cleavage
3.4. Cell Culture
3.5. MTT Cell Cytotoxicity Assay
3.6. Time and Concentration Dependence of DNA Cleavage
3.7. DNA Cleavage Protection Assays
3.8. Assessment of DNA Cleavage under Hypoxic Conditions
Acknowledgements
- Samples Availability: Available from the authors.
References and Notes
- Brahic, C; Darro, F; Belloir, M; Bastide, J; Kiss, R; Delfourne, E. Synthesis and cytotoxic evaluation of analogues of the marine pyridoacridine amphimedine. Bioorg Med Chem 2002, 10, 2845–2853. [Google Scholar] [CrossRef] [PubMed]
- Delfourne, E; Darro, F; Bontemps-Subielos, N; Decaestecker, C; Bastide, J; Frydman, A; Kiss, R. Synthesis and characterization of the antitumor activities of analogues of meridine, a marine pyridoacridine alkaloid. J Med Chem 2001, 44, 3275–3282. [Google Scholar] [CrossRef] [PubMed]
- Delfourne, E; Darro, F; Portefaix, P; Galaup, C; Bayssade, S; Bouteille, A; Le Corre, L; Bastide, J; Collignon, F; Lesur, B; Frydman, A; Kiss, R. Synthesis and in vitro antitumor activity of novel ring D analogues of the marine pyridoacridine ascididemin: structure-activity relationship. J Med Chem 2002, 45, 3765–3771. [Google Scholar] [CrossRef] [PubMed]
- Delfourne, E; Kiss, R; Le Corre, L; Dujols, F; Bastide, J; Collignon, F; Lesur, B; Frydman, A; Darro, F. Synthesis and in vitro antitumor activity of phenanthrolin-7-one derivatives, analogues of the marine pyridoacridine alkaloids ascididemin and meridine: structure-activity relationship. J Med Chem 2003, 46, 3536–3545. [Google Scholar] [CrossRef] [PubMed]
- Delfourne, E; Kiss, R; Le Corre, L; Dujols, F; Bastide, J; Collignon, F; Lesur, B; Frydman, A; Darro, F. Synthesis and in vitro antitumor activity of ring C and D-substituted phenanthrolin-7-one derivatives, analogues of the marine pyridoacridine alkaloids ascididemin and meridine. Bioorg Med Chem 2004, 12, 3987–3994. [Google Scholar] [CrossRef] [PubMed]
- Delfourne, E; Kiss, R; Le Corre, L; Merza, J; Bastide, J; Frydman, A; Darro, F. Synthesis and in vitro antitumor activity of an isomer of the marine pyridoacridine alkaloid ascididemin and related compounds. Bioorg Med Chem 2003, 11, 4351–4356. [Google Scholar] [CrossRef] [PubMed]
- Feliu, L; Vera-Luque, P; Albericio, F; Alvarez, M. Advances in solid-phase cycloadditions for heterocyclic synthesis. J Comb Chem 2007, 9, 521–565. [Google Scholar] [CrossRef] [PubMed]
- LaBarbera, DV; Bugni, TS; Ireland, CM. The total synthesis of neoamphimedine. J Org Chem 2007, 72, 8501–8505. [Google Scholar] [CrossRef] [PubMed]
- Marshall, KM; Barrows, LR. Biological activities of pyridoacridines. Nat Prod Rep 2004, 21, 731–751. [Google Scholar] [CrossRef] [PubMed]
- Marshall, KM; Matsumoto, SS; Holden, JA; Concepcion, GP; Tasdemir, D; Ireland, CM; Barrows, LR. The anti-neoplastic and novel topoisomerase II-mediated cytotoxicity of neoamphimedine, a marine pyridoacridine. Biochem Pharmacol 2003, 66, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Tasdemir, D; Marshall, KM; Mangalindan, GC; Concepcion, GP; Barrows, LR; Harper, MK; Ireland, CM. Deoxyamphimedine, a new pyridoacridine alkaloid from two tropical Xestospongia sponges. J Org Chem 2001, 66, 3246–3248. [Google Scholar] [CrossRef] [PubMed]
- Fridovich, I. Biological effects of the superoxide radical. Arch Biochem Biophys 1986, 247, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B; Gutteridge, JMC. Free Radicals in Biology and Medicine, 2nd ed; Clarendon Press: Oxford, USA, 1989. [Google Scholar]
- Spencer, JP; Jenner, A; Aruoma, OI; Cross, CE; Wu, R; Halliwell, B. Oxidative DNA damage in human respiratory tract epithelial cells. Time course in relation to DNA strand breakage. Biochem Biophys Res Commun 1996, 224, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Sarker, AH; Watanabe, S; Seki, S; Akiyama, T; Okada, S. Oxygen radical-induced single-strand DNA breaks and repair of the damage in a cell-free system. Mutat Res 1995, 337, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Ferri, E; Gattavecchia, E; Feroci, G; Battino, M. Interaction between reactive oxygen species and coenzyme Q10 in an aprotic medium: a cyclic voltammetry study. Mol Aspects Med 1994, 15(Suppl), s83–s88. [Google Scholar] [PubMed]
- Crawford, PW; Gross, J; Lawson, K; Cheng, CC; Dong, Q; Liu, DF; Luo, YL; Szczepankiewicz, BG; Heathcock, CH. Electrochemical properties of some biologically active quinone derivatives: furanquinones, pyridoquinones, and diplamine, a cytotoxic pyridoacridine alkaloid. J Electrochem Soc 1997, 144, 3710–3715. [Google Scholar]
- Malins, DC; Holmes, EH; Polissar, NL; Gunselman, SJ. The etiology of breast cancer. Characteristic alteration in hydroxyl radical-induced DNA base lesions during oncogenesis with potential for evaluating incidence risk. Cancer 1993, 71, 3036–3043. [Google Scholar] [PubMed]
- Cortes, F; Pinero, J; Palitti, F. Cytogenetic effects of inhibition of topoisomerase I or II activities in the CHO mutant EM9 and its parental line AA8. Mutat Res 1993, 288, 281–289. [Google Scholar] [PubMed]
- Cantoni, O; Murray, D; Meyn, RE. Induction and repair of DNA single-strand breaks in EM9 mutant CHO cells treated with hydrogen peroxide. Chem Biol Interact 1987, 63, 29–38. [Google Scholar] [PubMed]
- Messer, J; Reynolds, M; Stoddard, L; Zhitkovich, A. Causes of DNA single-strand breaks during reduction of chromate by glutathione in vitro and in cells. Free Radic Biol Med 2006, 40, 1981–1992. [Google Scholar] [PubMed]
- Jeggo, PA; Caldecott, K; Pidsley, S; Banks, GR. Sensitivity of Chinese hamster ovary mutants defective in DNA double strand break repair to topoisomerase II inhibitors. Cancer Res 1989, 49, 7057–7063. [Google Scholar] [PubMed]
- Ding, Q; Chichak, K; Lown, JW. Pyrroloquinoline and pyridoacridine alkaloids from marine sources. Curr Med Chem 1999, 6, 1–27. [Google Scholar] [PubMed]
- Matsumoto, SS; Biggs, J; Copp, BR; Holden, JA; Barrows, LR. Mechanism of ascididemin-induced cytotoxicity. Chem Res Toxicol 2003, 16, 113–122. [Google Scholar] [PubMed]
- McDonald, LA; Eldredge, GS; Barrows, LR; Ireland, CM. Inhibition of topoisomerase II catalytic activity by pyridoacridine alkaloids from a Cystodytes sp. ascidian: a mechanism for the apparent intercalator-induced inhibition of topoisomerase II. J Med Chem 1994, 37, 3819–3827. [Google Scholar] [PubMed]
- Matsumoto, SS; Sidford, MH; Holden, JA; Barrows, LR; Copp, BR. Mechanism of action studies of cytotoxic marine alkaloids: ascididemin exhibits thiol-dependent oxidative DNA cleavage. Tetrahedron Lett 2000, 41, 1667–1670. [Google Scholar]
- Herman, EH; Ferrans, VJ. Reduction of chronic doxorubicin cardiotoxicity in dogs by pretreatment with (+/−)-1,2-bis(3,5-dioxopiperazinyl-1-yl)propane (ICRF-187). Cancer Res 1981, 41, 3436–3440. [Google Scholar] [PubMed]
- Malins, DC. Identification of hydroxyl radical-induced lesions in DNA base structure: biomarkers with a putative link to cancer development. J Toxicol Environ Health 1993, 40, 247–261. [Google Scholar] [PubMed]
- Doroshow, JH. Role of hydrogen peroxide and hydroxyl radical formation in the killing of Ehrlich tumor cells by anticancer quinones. Proc Natl Acad Sci USA 1986, 83, 4514–4518. [Google Scholar] [PubMed]
- Doroshow, JH; Davies, KJ. Redox cycling of anthracyclines by cardiac mitochondria. II. Formation of superoxide anion, hydrogen peroxide, and hydroxyl radical. J Biol Chem 1986, 261, 3068–3074. [Google Scholar] [PubMed]
- Doroshow, JH; Locker, GY; Ifrim, I; Myers, CE. Prevention of doxorubicin cardiac toxicity in the mouse by N-acetylcysteine. J Clin Invest 1981, 68, 1053–1064. [Google Scholar] [PubMed]
- Doroshow, JH; Locker, GY; Myers, CE. Enzymatic defenses of the mouse heart against reactive oxygen metabolites: alterations produced by doxorubicin. J Clin Invest 1980, 65, 128–135. [Google Scholar] [PubMed]
- Herman, EH; Ferrans, VJ; Jordan, W; Ardalan, B. Reduction of chronic daunorubicin cardiotoxicity by ICRF-187 in rabbits. Res Commun Chem Pathol Pharmacol 1981, 31, 85–97. [Google Scholar] [PubMed]
- de Guzman, FS; Carte, B; Troupe, N; Faulkner, DJ; Harper, MK; Concepcion, GP; Mangalindan, GC; Matsumoto, SS; Barrows, LR; Ireland, CM. Neoamphimedine: a new pyridoacridine topoisomerase II inhibitor which catenates DNA. J Org Chem 1999, 64 , 1400–1402. [Google Scholar] [PubMed]
- Englund, PT. The replication of kinetoplast DNA networks in Crithidia fasciculata. Cell 1978, 14, 157–168. [Google Scholar] [PubMed]
- Matsumoto, SS; Haughey, HM; Schmehl, DM; Venables, DA; Ireland, CM; Holden, JA; Barrows, LR. Makaluvamines vary in ability to induce dose-dependent DNA cleavage via topoisomerase II interaction. Anticancer Drugs 1999, 10, 39–45. [Google Scholar] [PubMed]
- Holden, JA; Rolfson, DH; Low, RL. DNA topoisomerase I from human placenta. Biochim. Biophys. Acta 1990, 1049, 303–310. [Google Scholar] [PubMed]
- Mossman, T. Rapid colorimetric assay for cell growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983, 65, 55–63. [Google Scholar] [PubMed]
- Kokoshka, JM; Capson, TL; Holden, JA; Ireland, CM; Barrows, LR. Differences in the topoisomerase I cleavage complexes formed by camptothecin and wakayin, a DNA-intercalating marine natural product. Anticancer Drugs 1996, 7, 758–765. [Google Scholar] [PubMed]
- Bonnard, I; Bontemps, N; Lahmy, S; Banaigs, B; Combaut, G; Francisco, C; Colson, P; Houssier, C; Waring, MJ; Bailly, C. Binding to DNA and cytotoxic evaluation of ascididemin, the major alkaloid from the Mediterranean ascidian Cystodytes dellechiajei. Anticancer Drug Des 1995, 10, 333–346. [Google Scholar] [PubMed]
- Radisky, DC; Radisky, E; Barrows, LR; Copp, BR; Kramer, RA; Ireland, CM. Novel cytotoxic topoisomerase II inhibiting pyrroloiminoquinones from Fijian sponges of the Genus Zyzzya. J Am Chem Soc 1993, 115, 1632–1638. [Google Scholar]



| Cultured Mammalian Cell Line | IC50 in μM |
|---|---|
| Human melanoma, SK-mel-5 | 5.5 |
| Human epidermoid-nasopharyngeal cancer, KB | 3.8 |
| Human breast cancer, MCF7 | 5.9 |
| Human ovarian cancer-wild type, A2780wt | 0.3 |
| Human ovarian cancer-multi-drug resistant, A2780AD | 0.4 |
| Chinese hamster ovary, CHO wild type, AA8 | 13.7 |
| CHO-double strand (ds) DNA break repair deficient, xrs-6 | 4.2 |
| CHO-single strand (ss) DNA break repair deficient, EM9 | 3.8 |
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Marshall, K.M.; Andjelic, C.D.; Tasdemir, D.; Concepción, G.P.; Ireland, C.M.; Barrows, L.R. Deoxyamphimedine, a Pyridoacridine Alkaloid, Damages DNA via the Production of Reactive Oxygen Species. Mar. Drugs 2009, 7, 196-209. https://doi.org/10.3390/md7020196
Marshall KM, Andjelic CD, Tasdemir D, Concepción GP, Ireland CM, Barrows LR. Deoxyamphimedine, a Pyridoacridine Alkaloid, Damages DNA via the Production of Reactive Oxygen Species. Marine Drugs. 2009; 7(2):196-209. https://doi.org/10.3390/md7020196
Chicago/Turabian StyleMarshall, Kathryn M., Cynthia D. Andjelic, Deniz Tasdemir, Gisela P. Concepción, Chris M. Ireland, and Louis R. Barrows. 2009. "Deoxyamphimedine, a Pyridoacridine Alkaloid, Damages DNA via the Production of Reactive Oxygen Species" Marine Drugs 7, no. 2: 196-209. https://doi.org/10.3390/md7020196




