Natural Occurrence of 2′,5′-Linked Heteronucleotides in Marine Sponges
Abstract
:1. Introduction
Results and Discussion
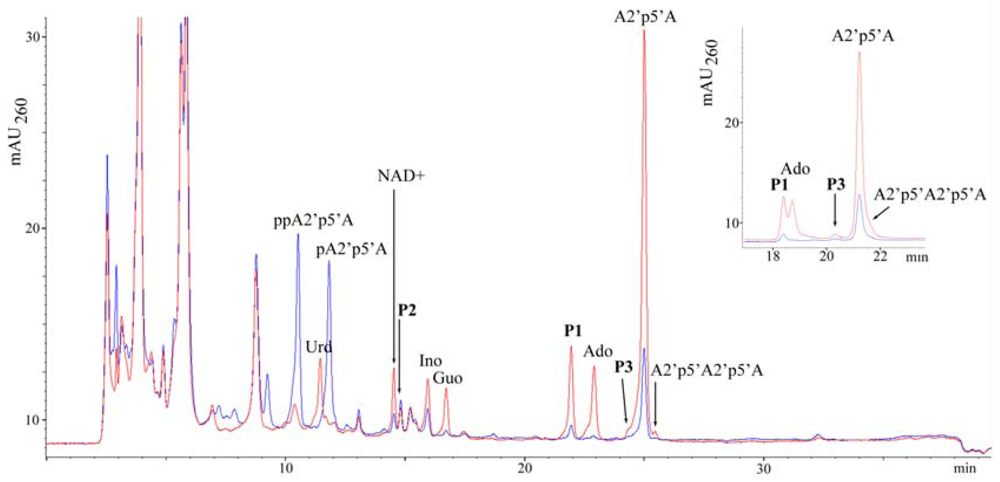
2.1. Analysis of sponge perchloric acid extracts
2.2. The formation of 2′,5′-linked oligomers in vitro
3. Experimental Section
3.1. Sponges
3.2. Sample homogenization
3.3. Preparation of extracts
3.3.1. Perchloric acid extract
3.3.2. Protein extract
3.4. 2′,5′-Oligoadenylate synthetase activity assays
3.5. HPLC analysis
3.6. Analysis of the composition of oligoribonucleotides by means of chemical and enzymatic hydrolysis
3.6.1. Alkaline hydrolysis
3.6.2. Snake venom phosphodiesterase treatment
3.6.3. Ribonuclease T2 treatment
3.7. MALDI-MS analysis
4. Conclusions
Acknowledgements
References
- Hovanessian, AG. On the discovery of interferon-inducible, double-stranded RNA activated enzymes: the 2′–5′oligoadenylate synthetases and the protein kinase PKR. Cytokine Growth Factor Rev 2007, 18, 351–361. [Google Scholar]
- Williams, BR; Golgher, RR; Kerr, IM. Activation of a nuclease by pppA2′p5′A2′p5′A in intact cells. FEBS Lett 1979, 105, 47–52. [Google Scholar]
- Hovanessian, AG; Wood, JN. Anticellular and antiviral effects of pppA(2′p5′A)n. Virology 1980, 101, 81–90. [Google Scholar]
- Zhou, A; Hassel, BA; Silverman, RH. Expression cloning of 2-5A-dependent RNAase: A uniquely regulated mediator of interferon action. Cell 1993, 72, 753–765. [Google Scholar]
- Kerr, IM. The 2–5A system: A personal view. J Interferon Res 1987, 7, 505–510. [Google Scholar]
- Salzberg, S; Hyman, T; Turm, H; Kinar, Y; Schwartz, Y; Nir, U; Lejbkowicz, F; Huberman, E. Ectopic expression of 2-5A synthetase in myeloid cells induces growth arrest and facilitates the appearance of a myeloid differentiation marker. Cancer Res 1997, 57, 2732–2740. [Google Scholar]
- Player, MR; Torrence, PF. The 2-5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol Ther 1998, 78, 55–113. [Google Scholar]
- Ghosh, A; Sarkar, SN; Sen, GC. Cell growth regulatory and antiviral effects of the P69 isozyme of 2-5 (A) synthetase. Virology 2000, 266, 319–328. [Google Scholar]
- Chawla-Sarkar, M; Lindner, DJ; Liu, YF; Williams, BR; Sen, GC; Silverman, RH; Borden, EC. Apoptosis and interferons: Role of interferon-stimulated genes as mediators of apoptosis. Apoptosis 2003, 8, 237–249. [Google Scholar]
- Ghosh, A; Sarkar, SN; Rowe, TM; Sen, GC. A specific isozyme of 2′–5′ oligoadenylate synthetase is a dual function proapoptotic protein of the Bcl-2 family. J Biol Chem 2001, 276, 25447–25455. [Google Scholar]
- McAveney, KM; Book, ML; Ling, P; Chebath, J; Yu-Lee, L. Association of 2′,5′-oligoadenylate synthetase with the prolactin (PRL) receptor: alteration in PRL-inducible stat1 (signal transducer and activator of transcription 1) signaling to the IRF-1 (interferonregulatory factor 1) promoter. Mol Endocrinol 2000, 14, 295–306. [Google Scholar]
- Castora, FJ; Erickson, CE; Kovács, T; Lesiak, K; Torrence, PF. 2′,5′-Oligoadenylates inhibit relaxation of supercoiled DNA by calf thymus DNA topoisomerase I. J Interferon Res 1991, 11, 143–149. [Google Scholar]
- Justesen, J; Ferbus, D; Thang, MN. Elongation mechanism and substrate specificity of 2′,5′-oligoadenylate synthetase. Proc Natl Acad Sci USA 1980, 77, 4618–4622. [Google Scholar]
- Justesen, J; Ferbus, D; Thang, MN. 2′5′ oligoadenylate synthetase, an interferon induced enzyme: direct assay methods for the products, 2′5′ oligoadenylates and 2′5′ co-oligonucleotides. Nucleic Acids Res 1980, 8, 3073–3085. [Google Scholar]
- Ferbus, D; Justesen, J; Besançon, F; Thang, MN. The 2′5′ oligoadenylate synthetase has a multifunctional 2′5′ nucleotidyl-transferase activity. Biochem Biophys Res Commun 1981, 100, 847–856. [Google Scholar]
- Marié, I; Blanco, J; Rebouillat, D; Hovanessian, AG. 69-kDa and 100-kDa isoforms of interferon-induced (2′–5′)oligoadenylate synthetase exhibit differential catalytic parameters. Eur J Biochem 1997, 248, 558–566. [Google Scholar]
- Voet, D; Voet, JG. Biochemistry; J. Wiley and Sons: New York, NY, USA, 1990. [Google Scholar]
- Sperling, J; Chebath, J; Arad-Dann, H; Offen, D; Spann, P; Lehrer, R; Goldblatt, D; Jolles, B; Sperling, R. Possible involvement of (2′–5′)oligoadenylate synthetase activity in pre-mRNA splicing. Proc Natl Acad Sci USA 1991, 88, 10377–10381. [Google Scholar]
- Besse, S; Rebouillat, D; Marie, I; Puvion-Dutilleul, F; Hovanessian, AG. Ultrastructural localization of interferon-inducible double-stranded RNA-activated enzymes in human cells. Exp Cell Res 1998, 239, 379–392. [Google Scholar]
- Drocourt, JL; Dieffenbach, CW; Ts’o, PO; Justesen, J; Thang, MN. Structural requirements of (2′–5′) oligoadenylate for protein synthesis inhibition in human fibroblasts. Nucleic Acids Res 1982, 10, 2163–2174. [Google Scholar]
- Justesen, J; Hartmann, R; Kjeldgaard, NO. Gene structure and function of the 2′–5′-oligoadenylate synthetase family. Cell Mol Life Sci 2000, 57, 1593–1612. [Google Scholar]
- Holm, L; Sander, C. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochim Sci 1995, 20, 345–347. [Google Scholar]
- Aravind, A; Koonin, E. DNA polymerase β-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res 1999, 27, 1609–1618. [Google Scholar]
- Martin, G; Keller, W. RNA-specific ribonucleotidyltransferases. RNA 2007, 11, 1834–1849. [Google Scholar]
- Torralba, S; Sojat, J; Hartmann, R. 2′–5′ oligoadenylate synthetase shares active site architecture with the archaeal CCA-adding enzyme. Cell Mol Life Sci 2008, 65, 2613–2620. [Google Scholar]
- Kjaer, KH; Poulsen, JB; Reintamm, T; Saby, E; Martensen, PM; Kelve, M; Justesen, J. Evolution of the 2′–5′-Oligoadenylate Synthetase Family in Eukaryotes and Bacteria. J Mol Evol 2009, 69, 612–624. [Google Scholar]
- Mashimo, T; Glaser, P; Lucas, M; Simon-Chazottes, D; Ceccaldi, PE; Montagutelli, X; Desprès, P; Guénet, JL. Structural and functional genomics and evolutionary relationships in the cluster of genes encoding murine 2′,5′-oligoadenylate synthetases. Genomics 2003, 82, 537–552. [Google Scholar]
- Venkatesh, B; Kirkness, EF; Loh, YH; Halpern, AL; Lee, AP; Johnson, J; Dandona, N; Viswanathan, LD; Tay, A; Venter, JC; Strausberg, RL; Brenner, S. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol 2007, 5, e101. [Google Scholar]
- Kuusksalu, A; Pihlak, A; Müller, WE; Kelve, M. The (2′–5′)oligoadenylate synthetase is present in the lowest multicellular organisms, the marine sponges. Demonstration of the existence and identification of its reaction products. Eur J Biochem 1995, 232, 351–357. [Google Scholar]
- Kuusksalu, A; Subbi, J; Pehk, T; Reintamm, T; Müller, WE; Kelve, M. Identification of the reaction products of (2′-5′)oligoadenylate synthetase in the marine sponge. Eur J Biochem 1998, 257, 420–426. [Google Scholar]
- Wiens, M; Kuusksalu, A; Kelve, M; Müller, WE. Origin of the interferon-inducible (2′–5′)oligoadenylate synthetases: Cloning of the (2′–5′)oligoadenylate synthetase from the marine sponge Geodia cydonium. FEBS Lett 1999, 462, 12–18. [Google Scholar]
- Grebenjuk, VA; Kuusksalu, A; Kelve, M; Schütze, J; Schröder, HC; Müller, WE. Induction of (2′–5′)oligoadenylate synthetase in the marine sponges Suberites domuncula and Geodia cydonium by the bacterial endotoxin lipopolysaccharide. Eur J Biochem 2002, 269, 1382–1392. [Google Scholar]
- Müller, WEG; Müller, IM. Origin of the metazoan immune system: Identification of the molecules and their functions in sponges. Integr Comp Biol 2003, 43, 281–292. [Google Scholar]
- Schröder, HC; Natalio, F; Wiens, M; Tahir, MN; Shukoor, MI; Tremel, W; Belikov, SI; Krasko, A; Müller, WEG. The 2′–5′-oligoadenylate in the lowest metazoa: isolation, cloning, expression and functional activity in the sponge Lubomirskia baicalensis. Mol Immunol 2008, 45, 945–953. [Google Scholar]
- Päri, M; Kuusksalu, A; Lopp, A; Reintamm, T; Justesen, J; Kelve, M. Expression and characterization of recombinant 2′,5′-oligoadenylate synthetase from the marine sponge Geodia cydonium. FEBS J 2007, 274, 3462–3474. [Google Scholar]
- Reintamm, T; Kuusksalu, A; Metsis, M; Päri, M; Vallmann, K; Lopp, A; Justesen, J; Kelve, M. Sponge OAS has a distinct genomic structure within the 2-5A synthetase family. Mol Genet Genomics 2008, 280, 453–466. [Google Scholar]
- Schröder, HC; Wiens, M; Kuusksalu, A; Kelve, M; Müller, WEG. Modulation of 2′-5′ oligoadenylate synthetase by environmental stress in marine sponge Geodia cydonium. Environ Toxicol Chem 1997, 16, 1403–1409. [Google Scholar]
- Robalino, J; Bartlett, TC; Chapman, RW; Gross, PS; Browdy, CL; Warr, GW. Doublestranded RNA and antiviral immunity in marine shrimp: inducible host mechanisms and evidence for the evolution of viral counter-responses. Dev Comp Immunol 2007, 31, 539–547. [Google Scholar]
- Reintamm, T; Lopp, A; Kuusksalu, A; Subbi, J; Kelve, M. Qualitative and quantitative aspects of 2-5A synthesizing capacity of different marine sponges. Biomol Eng 2003, 20, 389–399. [Google Scholar]
- Wells, JA; Swyryd, EA; Stark, GR. An improved method for purifying 2′,5′-oligoadenylate synthetases. J Biol Chem 1984, 259, 1363–1370. [Google Scholar]
- Amantonico, A; Urban, PL; Zenobi, R. Facile analysis of metabolites by capillary electrophoresis coupled to matrix-assisted laser desorption/ionization mass spectrometry using target plates with polysilazane nanocoating and grooves. Analyst 2009, 134, 1536–1540. [Google Scholar]
- Cayley, PJ; Kerr, IM. Synthesis, characterisation and biological significance of (2′-5′)oligoadenylate derivatives of NAD+, ADP-ribose and adenosine(5′)tetraphospho(5′)adenosine. Eur J Biochem 1982, 122, 601–608. [Google Scholar]
- Seth, M; Thurlow, DL; Hou, YM. Poly(C) synthesis by class I and class II CCA-adding enzymes. Biochemistry 2002, 41, 4521–4532. [Google Scholar]
- Bradford, MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976, 72, 248–254. [Google Scholar]
- Lopp, A; Kuusksalu, A; Reintamm, T; Müller, WEG; Kelve, M. 2′,5′-oligoadenylate synthetase from a lower invertebrate, the marine sponge Geodia cydonium, does not need dsRNA for its enzymatic activity. Biochim Biophys Acta 2002, 1590, 140–149. [Google Scholar]






| Substrate | (2′–5′) Oligomer*1 | Retention time, min*2 | m/zexp*3 |
|---|---|---|---|
| ATP | ApA | 22.2 | 595.1 |
| ApApA | 22.5 | 924.1 | |
| ApApApA | 21.9 | 1253.1 | |
| ApApApApA | 21.0 | 1582.3 | |
| GTP | GpG | 14.9 | 627.1 |
| GpGpG | 17.5 | 972.1 | |
| GpGpGpG | 17.9 | 1317.3 | |
| UTP | UpU | 15.1 | 549.0 |
| UpUpU | 16.3 | 855.1 | |
| CTP | CpC | 10.6 | 547.0 |
| CpCpC | 11.4 | 852.0 | |
| Thenea muricata | ||||||
|---|---|---|---|---|---|---|
| Substrate | Incubation time, h | Monomer* | Dimer | Trimer | Tetramer | Pentamer |
| ATP | 17 | 3.4 | 27.6 | 48.7 | 18.5 | 1.9 |
| GTP | 20 | 5.7 | 67 | 24.1 | 3.3 | - |
| UTP | 20 | 5.1 | 85.6 | 9.3 | - | - |
| CTP | 20 | 7.4 | 88.1 | 4.5 | - | - |
| Chondrilla nucula | ||||||
| ATP | 1 | 4.1 | 31.3 | 59.6 | 5 | - |
| GTP | 1 | 10.1 | 50.6 | 35.7 | 3.6 | - |
| UTP | 18 | 13.6 | 76.4 | 10 | - | - |
| Substrates | (2′–5′) Oligomer*1 | Retention time, min*2 | m/zexp*3 | Sponge |
|---|---|---|---|---|
| ATP + GTP | GpGpA | 16.7 | 956.1 | 2 |
| GpGpApA | 16.9 | 1285.2 | 2 | |
| GpA | 17.5 | 611.1 | 1, 2 | |
| GpApA | 18.1 | 940.1 | 1, 2 | |
| ApGpG | 19.2 | 956.1 | 2 | |
| ApG | 19.4 | 611.1 | 1, 2 | |
| ApApG | 20.2 | 940.2 | 1, 2 | |
| GpApG | 20.3 | 956.2 | 2 | |
| ApGpA | 20.7 | 940.2 | 2 | |
| ATP + UTP | UpApU | 17.7 | 878.0 | 1 |
| UpApApA | 18.4 | 1230.1 | 1 | |
| UpA | 18.8 | 572.0 | 1 | |
| UpApA | 19.1 | 901.0 | 1 | |
| ApApU | 19.9 | 901.0 | 1, 2 | |
| ApApApU | 19.9 | 1230.1 | 1 | |
| ApU | 20.9 | 572.0 | 1, 2 | |
| ATP + CTP | CpApC | 13.7 | 876.1 | 1 |
| CpA | 15.5 | 571.1 | 1 | |
| CpApA | 15.5 | 900.1 | 1 | |
| ApApApC | 19.5 | 1229.2 | 1 | |
| ApApU*4 | 19.9 | 901.1 | 2 | |
| ApApC | 19.9 | 900.1 | 1, 2 | |
| ApU*4 | 20.9 | 572.1 | 2 | |
| ApC | 21.0 | 571.1 | 1, 2 | |
| GTP + UTP | GpGpU | 14.6 | 933.1 | 2 |
| GpGpGpU | 15.7 | 1278.1 | 2 | |
| GpU | 16.0 | 588.1 | 2 | |
| ATP + NAD+ | NADpA | 13.2 | 993.1*5 | 1, 2 |
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lopp, A.; Reintamm, T.; Kuusksalu, A.; Tammiste, I.; Pihlak, A.; Kelve, M. Natural Occurrence of 2′,5′-Linked Heteronucleotides in Marine Sponges. Mar. Drugs 2010, 8, 235-254. https://doi.org/10.3390/md8020235
Lopp A, Reintamm T, Kuusksalu A, Tammiste I, Pihlak A, Kelve M. Natural Occurrence of 2′,5′-Linked Heteronucleotides in Marine Sponges. Marine Drugs. 2010; 8(2):235-254. https://doi.org/10.3390/md8020235
Chicago/Turabian StyleLopp, Annika, Tönu Reintamm, Anne Kuusksalu, Indrek Tammiste, Arno Pihlak, and Merike Kelve. 2010. "Natural Occurrence of 2′,5′-Linked Heteronucleotides in Marine Sponges" Marine Drugs 8, no. 2: 235-254. https://doi.org/10.3390/md8020235




