Synthesis and α-Glucosidase Inhibitory Mechanisms of Bis(2,3-dibromo-4,5-dihydroxybenzyl) Ether, a Potential Marine Bromophenol α-Glucosidase Inhibitor
Abstract
:1. Introduction
2. Results and Discussion
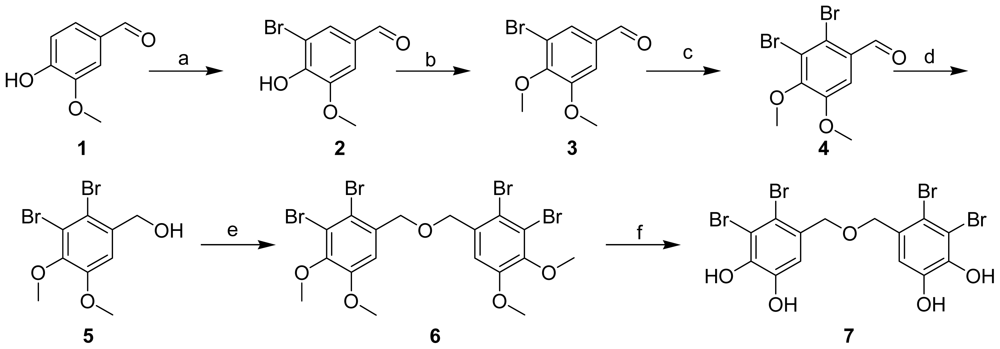
2.1. Synthesis of BDDE
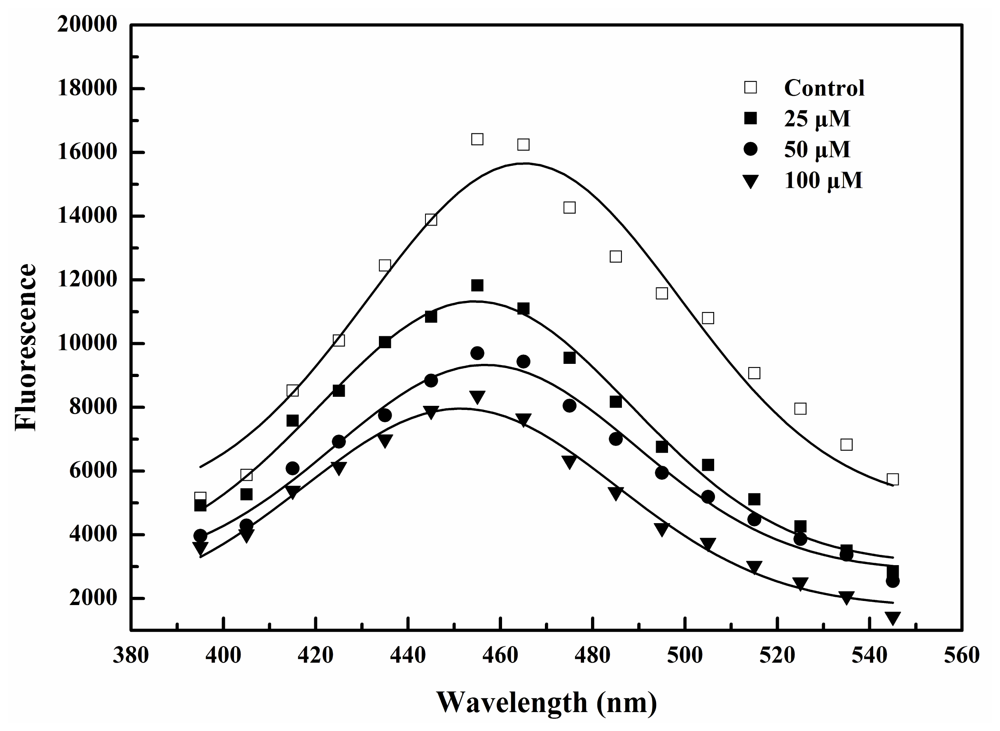
2.2. BDDE Binding Quenched the Intrinsic Fluorescence of α-Glucosidase
2.3. BDDE Binding Reduced the Hydrophobicity of α-Glucosidase
2.4. BDDE Binding Influenced the Secondary Structures of α-Glucosidase
2.5. Protein 3D Structure Generation and Molecular Docking
3. Experimental Section
3.1. Materials
3.2. Synthesis of BDDE
3-Bromo-4-hydroxy-5-methoxybenzaldehyde (2)
3-Bromo-4,5-dimethoxybenzaldehyde (3)
2,3-Dibromo-4,5-dimethoxybenzaldehyde (4)
(2,3-Dibromo-4,5-dimethoxyphenyl)methanol (5)
5,5′-Oxybis(methylene)bis(3,4-dibromo-1,2-dimethoxybenzene) (6)
Bis(2,3-dibromo-4,5-dihydroxybenzyl) ether (7)
3.3. Intrinsic Fluorescence Measurements
3.4. Hydrophobic Analysis of α-Glucosidase Using Bis-ANS
3.5. Circular Dichroism Spectroscopy
3.6. Molecular Modeling and Docking
4. Conclusions
Acknowledgments
- Samples Availability: Available from the authors.
References
- Lefebvre, PJ; Scheen, AJ. The use of acarbose in the prevention and treatment of hypoglycaemia. Eur. J. Clin. Invest 1994, 24(Suppl 3), 40–44. [Google Scholar]
- Scott, LJ; Spencer, CM. Miglitol: A review of its therapeutic potential in type 2 diabetes mellitus. Drugs 2000, 59, 521–549. [Google Scholar]
- Campbell, LK; Baker, DE; Campbell, RK. Miglitol: Assessment of its role in the treatment of patients with diabetes mellitus. Ann. Pharmacother 2000, 34, 1291–1301. [Google Scholar]
- Krentz, AJ; Bailey, CJ. Oral antidiabetic agents: current role in type 2 diabetes mellitus. Drugs 2005, 65, 385–411. [Google Scholar]
- Nathan, DM; Buse, JB; Davidson, MB; Heine, RJ; Holman, RR; Sherwin, R; Zinman, B. Management of hyperglycaemia in type 2 diabetes: A consensus algorithm for the initiation and adjustment of therapy. A consensus statement from the American Diabetes Association and the European Association for the Study of Diabetes. Diabetologia 2006, 49, 1711–1721. [Google Scholar]
- Hsiao, SH; Liao, LH; Cheng, PN; Wu, TJ. Hepatotoxicity associated with acarbose therapy. Ann. Pharmacother 2006, 40, 151–154. [Google Scholar]
- Du, ZY; Liu, RR; Shao, WY; Mao, XP; Ma, L; Gu, LQ; Huang, ZS; Chan, AS. Alpha-glucosidase inhibition of natural curcuminoids and curcumin analogs. Eur. J. Med. Chem 2006, 41, 213–218. [Google Scholar]
- de Melo, EB; da Silveira Gomes, A; Carvalho, I. alpha- and beta-glucosidase inhibitors: Chemical structure and biological activity. Tetrahedron 2006, 62, 10277–10302. [Google Scholar]
- Kwon, YI; Apostolidis, E; Shetty, K. In vitro studies of eggplant (Solanum melongena) phenolics as inhibitors of key enzymes relevant for type 2 diabetes and hypertension. Bioresour. Technol 2008, 99, 2981–2988. [Google Scholar]
- Tundis, R; Loizzo, MR; Menichini, F. Natural products as alpha-amylase and alpha-glucosidase inhibitors and their hypoglycaemic potential in the treatment of diabetes: An update. Mini Rev. Med. Chem 2010, 10, 315–331. [Google Scholar]
- Ranilla, LG; Kwon, Y-I; Apostolidis, E; Shetty, K. Phenolic compounds, antioxidant activity and in vitro inhibitory potential against key enzymes relevant for hyperglycemia and hypertension of commonly used medicinal plants, herbs and spices in Latin America. Bioresour. Technol 2010, 101, 4676–4689. [Google Scholar]
- Kurihara, H; Mitani, T; Kawabata, J; Takahashi, K. Two new bromophenols from the red alga odonthalia corymbifera. J. Nat. Prod 1999, 62, 882–884. [Google Scholar]
- Kurihara, H; Mitani, T; Kawabata, J; Takahashi, K. Inhibitory potencies of bromophenols from Rhodomelaceae algae against α-glucosidase activity. Fish Sci 1999, 65, 300–303. [Google Scholar]
- Kim, KY; Nam, KA; Kurihara, H; Kim, SM. Potent alpha-glucosidase inhibitors purified from the red alga Grateloupia elliptica. Phytochemistry 2008, 69, 2820–2825. [Google Scholar]
- Kim, KY; Nguyen, TH; Kurihara, H; Kim, SM. Alpha-glucosidase inhibitory activity of bromophenol purified from the red alga Polyopes lancifolia. J. Food. Sci 2010, 75, H145–H150. [Google Scholar]
- Xu, X; Song, F; Wang, S; Li, S; Xiao, F; Zhao, J; Yang, Y; Shang, S; Yang, L; Shi, J. Dibenzyl bromophenols with diverse dimerization patterns from the brown alga Leathesia nana. J. Nat. Prod 2004, 67, 1661–1666. [Google Scholar]
- Xu, N; Fan, X; Yan, X; Li, X; Niu, R; Tseng, CK. Antibacterial bromophenols from the marine red alga Rhodomela confervoides. Phytochemistry 2003, 62, 1221–1224. [Google Scholar]
- Oh, KB; Jeon, HB; Han, YR; Lee, YJ; Park, J; Lee, SH; Yang, D; Kwon, M; Shin, J; Lee, HS. Bromophenols as Candida albicans isocitrate lyase inhibitors. Bioorg. Med. Chem. Lett 2010, 20, 6644–6648. [Google Scholar]
- Yue, Q; Niu, L; Li, X; Shao, X; Xie, X; Song, Z. Study on the interaction mechanism of lysozyme and bromophenol blue by fluorescence spectroscopy. J. Fluoresc 2008, 18, 11–15. [Google Scholar]
- Qi, X; Grabowski, GA. Acid beta-glucosidase: intrinsic fluorescence and conformational changes induced by phospholipids and saposin C. Biochemistry 1998, 37, 11544–11554. [Google Scholar]
- Li, Y; Gao, F; Shan, F; Bian, J; Zhao, C. Study on the interaction between 3 flavonoid compounds and alpha-amylase by fluorescence spectroscopy and enzymatic kinetics. J. Food Sci 2009, 74, C199–C203. [Google Scholar]
- Hawe, A; Sutter, M; Jiskoot, W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res 2008, 25, 1487–1499. [Google Scholar]
- Liu, M; Zhang, W; Qiu, L; Lin, X. Synthesis of butyl-isobutyl-phthalate and its interaction with alpha-glucosidase in vitro. J. Biochem 2011, 149, 27–33. [Google Scholar]
- Kim, JH; Kim, HJ; Park, HW; Youn, SH; Choi, DY; Shin, CS. Development of inhibitors against lipase and alpha-glucosidase from derivatives of monascus pigment. FEMS Microbiol. Lett 2007, 276, 93–98. [Google Scholar]
- Shen, Q; Shao, J; Peng, Q; Zhang, W; Ma, L; Chan, AS; Gu, L. Hydroxycoumarin derivatives: Novel and potent alpha-glucosidase inhibitors. J. Med. Chem 2010, 53, 8252–8259. [Google Scholar]
- Yang, JT; Wu, CS; Martinez, HM. Calculation of protein conformation from circular dichroism. Methods Enzymol 1986, 130, 208–269. [Google Scholar]
- Price, MLP; Ostrovsky, D; Jorgensen, WL. Gas-phase and liquid-state properties of esters, nitriles, and nitro compounds with the OPLS-AA force field. J. Comput. Chem 2001, 22, 1340–1352. [Google Scholar]
- Sherman, W; Day, T; Jacobson, MP; Friesner, RA; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem 2006, 49, 534–553. [Google Scholar]




| Marine bromophenol | (μM) | Inhibition mode | Reference |
|---|---|---|---|
| Bis(2,3,6-tribromo-4,5-dihydroxybenzyl) ether | 0.03 | Unknown | [13] |
| Bis(2,3-dibromo-4,5-dihydroxybenzyl) ether | 0.098 | Competitive | [12,13,15] |
| 2,3,6-Tribromo-4,5-dihydroxybenzyl alcohol | 11 | Mixed | [15] |
| 4-Bromo-2,3-dihydroxy-6-hydroxymethylphenyl-2,5-dibromo-6-hydroxy-3-hydroxymethylphenyl ether | 25 | Unknown | [12] |
| 2,4,6-Tribromophenol | 60.3 | Mixed | [14] |
| 2,3-Dibromo-4,5-dihydroxybenzyl alcohol | 89 | Mixed | [12] |
| 3-Bromo-4,5-dihydroxybenzyl alcohol | 100 | Mixed | [13] |
| 2,4-Dibromophenol | 110.4 | Mixed | [14] |
| BDDE (μM) | α-Helix (%) | β-Sheet (%) | β-Turn (%) | Random (%) |
|---|---|---|---|---|
| 0 | 27.5 | 39.4 | 7.1 | 26 |
| 25 | 30.3 | 33.8 | 5.6 | 30.3 |
| 50 | 28.9 | 34.7 | 8.3 | 28.2 |
| 100 | 31.2 | 31.6 | 7.1 | 30.2 |
© 2011 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, M.; Zhang, W.; Wei, J.; Lin, X. Synthesis and α-Glucosidase Inhibitory Mechanisms of Bis(2,3-dibromo-4,5-dihydroxybenzyl) Ether, a Potential Marine Bromophenol α-Glucosidase Inhibitor. Mar. Drugs 2011, 9, 1554-1565. https://doi.org/10.3390/md9091554
Liu M, Zhang W, Wei J, Lin X. Synthesis and α-Glucosidase Inhibitory Mechanisms of Bis(2,3-dibromo-4,5-dihydroxybenzyl) Ether, a Potential Marine Bromophenol α-Glucosidase Inhibitor. Marine Drugs. 2011; 9(9):1554-1565. https://doi.org/10.3390/md9091554
Chicago/Turabian StyleLiu, Ming, Wei Zhang, Jianteng Wei, and Xiukun Lin. 2011. "Synthesis and α-Glucosidase Inhibitory Mechanisms of Bis(2,3-dibromo-4,5-dihydroxybenzyl) Ether, a Potential Marine Bromophenol α-Glucosidase Inhibitor" Marine Drugs 9, no. 9: 1554-1565. https://doi.org/10.3390/md9091554





