NMR-Based Metabolomic Investigations on the Differential Responses in Adductor Muscles from Two Pedigrees of Manila Clam Ruditapes philippinarum to Cadmium and Zinc
Abstract
:1. Introduction
2. Results and Discussion
2.1. Metabolic Differences in Adductor Muscles between White and Zebra Clams
2.2. Differential Responses in Adductor Muscles from White and Zebra Clams to Heavy Metal Exposures
3. Experimental
3.1. Chemicals
3.2. Clam Exposure
3.3. Metabolite Extraction
3.4. High Resolution One Dimensional 1H NMR Spectroscopy
3.5. Spectral Pre-Processing and Multivariate Data Analysis
4. Conclusions
Acknowledgments
- Samples Availability: Available from the authors.
References
- Daviss, B. Growing pains for metabolomics. Scientist 2005, 19, 25–28. [Google Scholar]
- Lindon, JC; Nicholson, JK; Holmes, E; Everett, JR. Metabonomics: Metabolic processes studied by NMR spectroscopy of biofluids. Concepts Magn. Reson 2000, 12, 289–320. [Google Scholar]
- Viant, MR; Eric, S; Rosenblum, ES; Tjeerdema, RS. Optimized method for the determination of phosphoarginine in abalone tissue by high-performance liquid chromatography. J. Chromatogr. B 2001, 765, 107–111. [Google Scholar]
- Brindle, JT; Antti, H; Holmes, E; Tranter, G; Nicholson, JK; Bethell, HWL; Clarke, S; Schofield, PM; McKilligin, E; Mosedale, DE; et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat. Med 2002, 8, 1439–1444. [Google Scholar]
- Bundy, JG; Spurgeon, DJ; Svendsen, C; Hankard, PK; Weeks, JM; Osborn, D; Lindon, JC; Nicholson, JK. Environmental metabonomics: Applying combination biomarker analysis in earthworms at a metal contaminated site. Ecotoxicology 2004, 13, 797–806. [Google Scholar]
- Wu, H; Zhang, X; Li, X; Wu, Y; Pei, F. Acute biochemical effects of La(NO3)3 on liver and kidney tissues by magic-angle spinning 1H nuclear magnetic resonance spectroscopy and pattern recognition. Anal. Biochem 2005, 339, 242–248. [Google Scholar]
- Viant, MR; Pincetich, CA; Hinton, DE; Tjeerdema, RS. Toxic actions of dinoseb in medaka (Oryzias latipes) embryos as determined by in vivo 31P NMR, HPLC-UV and 1H NMR metabolomics. Aquat. Toxicol 2006, 76, 329–342. [Google Scholar]
- Viant, MR; Pincetich, CA; Hinton, DE; Tjeerdema, RS. Metabolic effects of dinoseb, diazinon and esfenvalerate in eyed eggs and alevins of Chinook salmon (Oncorhynchus tshawytscha) determined by 1H NMR metabolomics. Aquat. Toxicol 2006, 77, 359–371. [Google Scholar]
- Nicholson, JK; Timbrell, JA; Sadler, PJ. Mercury nephrotoxicity and the detection of abnormal urinary metabolite excretion patterns by high resolution proton nuclear magnetic resonance spectroscopy. Mol. Pharmacol 1985, 27, 644–651. [Google Scholar]
- Plumb, RS; Stumpf, CL; Granger, JH; Castro-Perez, J; Haselden, JN; Dear, GJ. Use of liquid chromatography/time-of-flight mass spectrometry and multivariate statistical analysis shows promise for the detection of drug metabolites in biological fluids. Rapid Commun. Mass Spectrom 2003, 17, 2632–2638. [Google Scholar]
- Wang, Y; Bollard, ME; Keun, H; Antti, H; Beckonert, O; Ebbels, TM; Lindon, JC; Holmes, E; Tang, H; Nicholson, JK. Spectral editing and pattern recognition methods applied to high-resolution magic-angle spinning 1H nuclear magnetic resonance spectroscopy of liver tissues. Anal. Biochem 2003, 323, 26–32. [Google Scholar]
- Wu, H; Zhang, X; Wu, Y; Pei, F. Studies on the acute biochemical effects of La(NO3)3 using 1H NMR spectroscopy of urine combined with pattern recognition. J. Inorg. Biochem 2005, 99, 644–651. [Google Scholar]
- Jones, OAH; Dondero, F; Viarengo, A; Griffin, JL. Metabolic profiling of Mytilus galloprovincialis and its potential applications for pollution assessment. Mar. Ecol. Prog. Ser 2008, 369, 169–179. [Google Scholar]
- Tuffnail, W; Mills, GA; Cary, P; Greenwood, R. An environmental 1H NMR metabolomic study of the exposure of the marine mussel Mytilus edulis to atrazine, lindane, hypoxia and starvation. Metabolomics 2009, 5, 33–43. [Google Scholar]
- Wu, H; Wang, W-X. NMR-based metabolomic studies on the toxicological effects of cadmium and copper on green mussels Perna viridis. Aquat. Toxicol 2010, 100, 339–345. [Google Scholar]
- Tikunov, AP; Johnson, CB; Lee, H; Stoskopf, MK; Macdonald, JM. Metabolomic investigations of American oysters Using 1H-NMR spectroscopy. Mar. Drugs 2010, 8, 2578–2596. [Google Scholar]
- Lannig, G; Eilers, S; Pörtner, HO; Sokolova, IM; Bock, C. Impact of ocean acidification on energy metabolism of oyster, Crassostrea gigas—Changes in metabolic pathways and thermal response. Mar. Drugs 2010, 8, 2318–2339. [Google Scholar]
- Gordon, BR; Leggat, W. Symbiodinium—Invertebrate symbioses and the role of metabolomics. Mar. Drugs 2010, 8, 2546–2568. [Google Scholar]
- Ji, J; Choi, HJ; Ahn, I-Y. Evaluation of Manila clam Ruditapes philippinarum as a sentinel species for metal pollution monitoring in estuarine tidal flats of Korea: Effects of size, sex, and spawning on baseline accumulation. Mar. Pollut. Bull 2006, 52, 447–468. [Google Scholar]
- Laing, I; Child, AR. Comparative tolerance of small juvenile palourdes (Tapes decussates L.) and Manila clams (Tapes philippinarum Adams & Reeve) to low temperature. J. Exp. Mar. Biol. Ecol 1996, 195, 267–285. [Google Scholar]
- Matozzo, V; Ballarin, L; Marin, MG. Exposure of the clam Tapes philippinarum to 4-nonylphenol: Changes in anti-oxidant enzyme activities and re-burrowing capability. Mar. Pollut. Bull 2004, 48, 563–571. [Google Scholar]
- Hegaret, H; Da Silva, PM; Wikfors, GH; Lambert, C; de Bettignies, T; Shumway, SE; Soudant, P. Hemocyte responses of Manila clams, Ruditapes philippinarum, with varying parasite, Perkinsus olseni, severity to toxic-algal exposures. Aquat. Toxicol 2007, 84, 469–479. [Google Scholar]
- Moraga, D; Mdelgi-Lasram, E; Romdhane, MS; El Abed, A; Boutet, I; Tanguy, A; Auffret, M. Genetic responses to metal contamination in two clams: Ruditapes decussatus and Ruditapes philippinarum. Mar. Environ. Res 2002, 54, 521–525. [Google Scholar]
- Yan, X; Zhang, G; Yang, F; Liang, J. A comparison of growth and development of Manila Clam (Ruditapes philippinarum) from two pedigrees (in Chinese). J. Dalian Fish. Univ 2005, 20, 266–269. [Google Scholar]
- Zhang, Y; Yan, X; Yao, T; Huo, Z; Yang, F; Zhang, G. Study on population hybridization of two shell color strains of Manila clam Ruditapes philippinarum (in Chinese). South China Fish. Sci 2008, 4, 27–32. [Google Scholar]
- Liu, X; Zhang, L; You, L; Yu, J; Zhao, J; Li, L; Wang, Q; Li, F; Li, C; Liu, D; Wu, H. Differential toxicological effects induced by mercury in gills from three pedigrees of Manila clam Ruditapes philippinarum by NMR-based metabolomics. Ecotoxicology 2011, 20, 177–186. [Google Scholar]
- Mao, TY; Dai, MX; Peng, ST; Li, GL. Temporal-spatial variation trend analysis of heavy metals (Cu, Zn, Pb, Cd, Hg) in Bohai Bay in 10 Years (in Chinese). J. Tianjin Univ 2009, 9, 817–825. [Google Scholar]
- Zhang, X. Investigation of pollution of Pb, Cd, Hg, As in sea water and deposit of Bohai Sea area (in Chinese). Heilongjiang Environ. J 2001, 25, 87–90. [Google Scholar]
- Wu, H; Wang, W-X. Tissue-specific toxicological effects of cadmium in green mussel (Perna viridis): Nuclear magnetic resonance-based metabolomics study. Environ. Toxicol. Chem 2011, 30, 806–812. [Google Scholar]
- Wu, H; Southam, AD; Hines, A; Viant, MR. High throughput tissue extraction protocol for NMR and mass spectrometry based metabolomics. Anal. Biochem 2008, 372, 204–212. [Google Scholar]
- Lin, CY; Wu, H; Tjeerdema, RS; Viant, MR. Evaluation of metabolite extraction strategies from tissue samples using NMR metabolomics. Metabolomics 2007, 3, 55–67. [Google Scholar]
- Viant, MR; Rosenblum, ES; Tjeerdema, RS. NMR-based metabolomics: A powerful approach for characterizing the effects of environmental stressors on organism health. Environ. Sci. Technol 2003, 37, 4982–4989. [Google Scholar]
- Preston, RL. Transport of amino acids by marine invertebrates. Comp. Physiol. Biochem 2005, 265, 410–421. [Google Scholar]
- Liu, X; Zhang, L; You, L; Cong, M; Zhao, J; Wu, H; Li, C; Liu, D; Yu, J. Toxicological responses to acute mercury exposure for three species of Manila clam Ruditapes philippinarum by NMR-based metabolomics. Environ. Toxicol. Pharmacol 2011, 31, 323–332. [Google Scholar]
- Fan, WMT. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Prog. Nucl. Magn. Reson 1996, 28, 161–219. [Google Scholar]
- Purohit, PV; Rocke, DM; Viant, MR; Woodruff, DL. Discrimination models using variance-stabilizing transformation of metabolomic NMR data. OMICS J. Integr. Biol 2004, 8, 118–130. [Google Scholar]
- Parsons, HM; Ludwig, C; Gunther, UL; Viant, MR. Improved classification accuracy in 1- and 2-dimensional NMR metabolomics data using the variance stabilising generalised logarithm transformation. BMC Bioinform 2007, 8, 234. [Google Scholar]
- Xu, L. Methods of Chemometrics; Science Press: Beijing, China, 2004; pp. 221–227. [Google Scholar]
- Keun, HC; Ebbels, TMD; Antti, H; Bollard, ME; Beckonert, O; Holmes, E; Lindon, JC; Nicholson, JK. Improved analysis of multivariate data by variable stability scaling: Application to NMR-based metabolic profiling. Anal. Chim. Acta 2003, 490, 265–276. [Google Scholar]
- Rubingh, CM; Bijlsma, S; Derks, EPA; Bobeldijk, I; Verheij, ER; Kochhar, S; Smilde, AK. Assessing the performance of statistical validation tools for megavariate metabolomics data. Metabolomics 2006, 2, 53–61. [Google Scholar]
- Lindon, JC; Nicholson, JK; Everett, JR. NMR spectroscopy of biofluid. Ann. Rep. NMR Spectrosc 1999, 38, 1–88. [Google Scholar]
- Katsiadaki, I; Williams, TD; Ball, JS; Bean, TP; Sanders, MB; Wu, H; Santos, EM; Brown, MM; Baker, P; Ortega, F; et al. Hepatic transcriptomic and metabolomic responses in the Stickleback (Gasterosteus aculeatus) exposed to ethinyl-estradiol. Aquat. Toxicol 2009, 97, 174–187. [Google Scholar]

 ) and Zebra (
) and Zebra (
 ) clam samples, and corresponding LV1 (B) weights plot showing the metabolic differences between White and Zebra pedigrees of clam adductor muscle tissue extracts. Keys: (1) branched chain amino acids, (2) alanine, (3) unknown 1 (1.51 ppm), (4) arginine, (5) glutamine, (6) succinate, (7) aspartate, (8) hypotaurine, (9) 4-aminobutorate, (10) phosphocholine, (11) unknown 2 (3.72 ppm), (12) unknown 3 (4.12 ppm), (13) homarine and (14) histidine.
) clam samples, and corresponding LV1 (B) weights plot showing the metabolic differences between White and Zebra pedigrees of clam adductor muscle tissue extracts. Keys: (1) branched chain amino acids, (2) alanine, (3) unknown 1 (1.51 ppm), (4) arginine, (5) glutamine, (6) succinate, (7) aspartate, (8) hypotaurine, (9) 4-aminobutorate, (10) phosphocholine, (11) unknown 2 (3.72 ppm), (12) unknown 3 (4.12 ppm), (13) homarine and (14) histidine.
 ) and Zebra (
) and Zebra (
 ) clam samples, and corresponding LV1 (B) weights plot showing the metabolic differences between White and Zebra pedigrees of clam adductor muscle tissue extracts. Keys: (1) branched chain amino acids, (2) alanine, (3) unknown 1 (1.51 ppm), (4) arginine, (5) glutamine, (6) succinate, (7) aspartate, (8) hypotaurine, (9) 4-aminobutorate, (10) phosphocholine, (11) unknown 2 (3.72 ppm), (12) unknown 3 (4.12 ppm), (13) homarine and (14) histidine.
) clam samples, and corresponding LV1 (B) weights plot showing the metabolic differences between White and Zebra pedigrees of clam adductor muscle tissue extracts. Keys: (1) branched chain amino acids, (2) alanine, (3) unknown 1 (1.51 ppm), (4) arginine, (5) glutamine, (6) succinate, (7) aspartate, (8) hypotaurine, (9) 4-aminobutorate, (10) phosphocholine, (11) unknown 2 (3.72 ppm), (12) unknown 3 (4.12 ppm), (13) homarine and (14) histidine.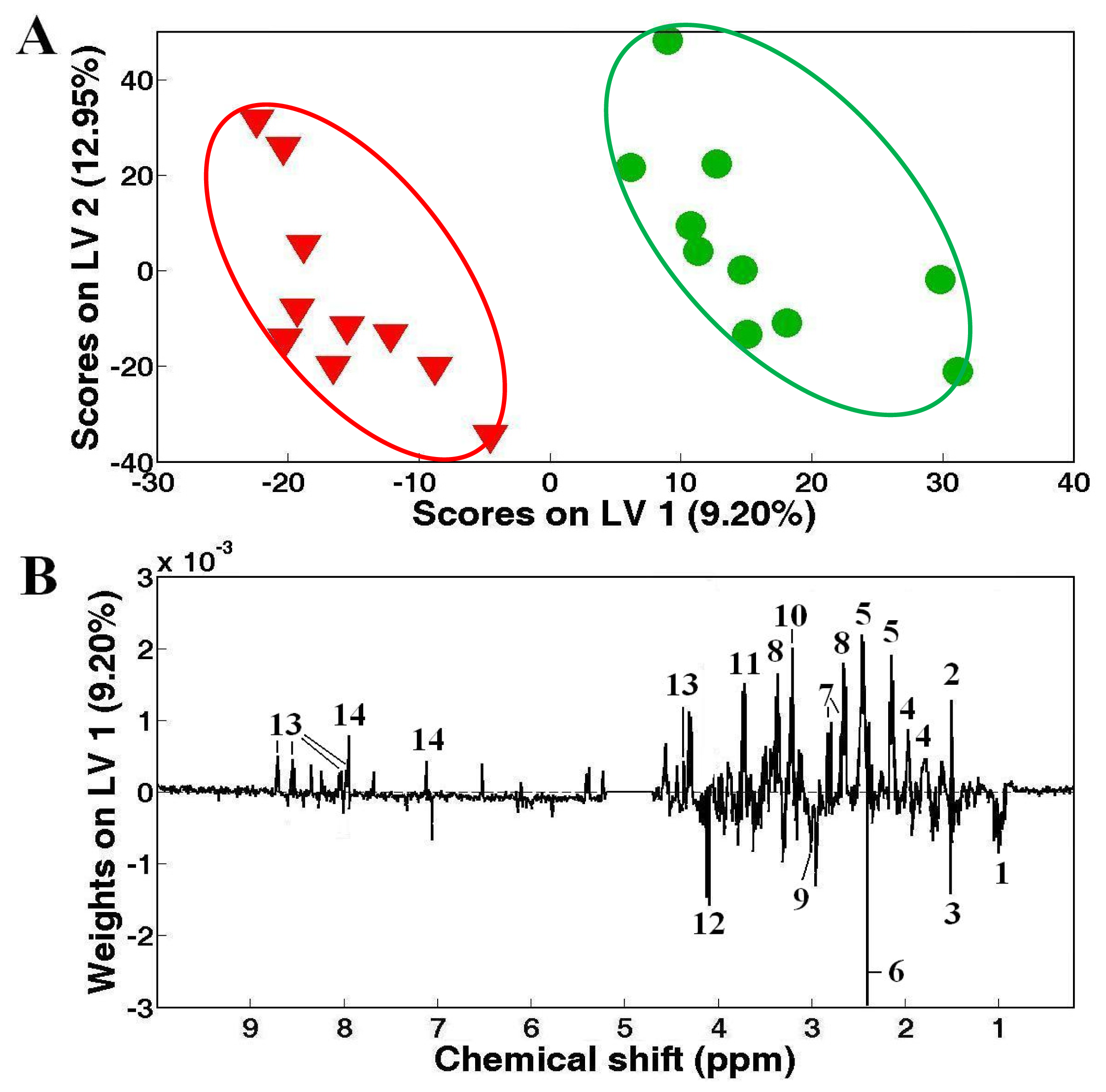
 ) and Cd (A), Zn (C) and mixed Cd and Zn (E) exposed (
) and Cd (A), Zn (C) and mixed Cd and Zn (E) exposed (
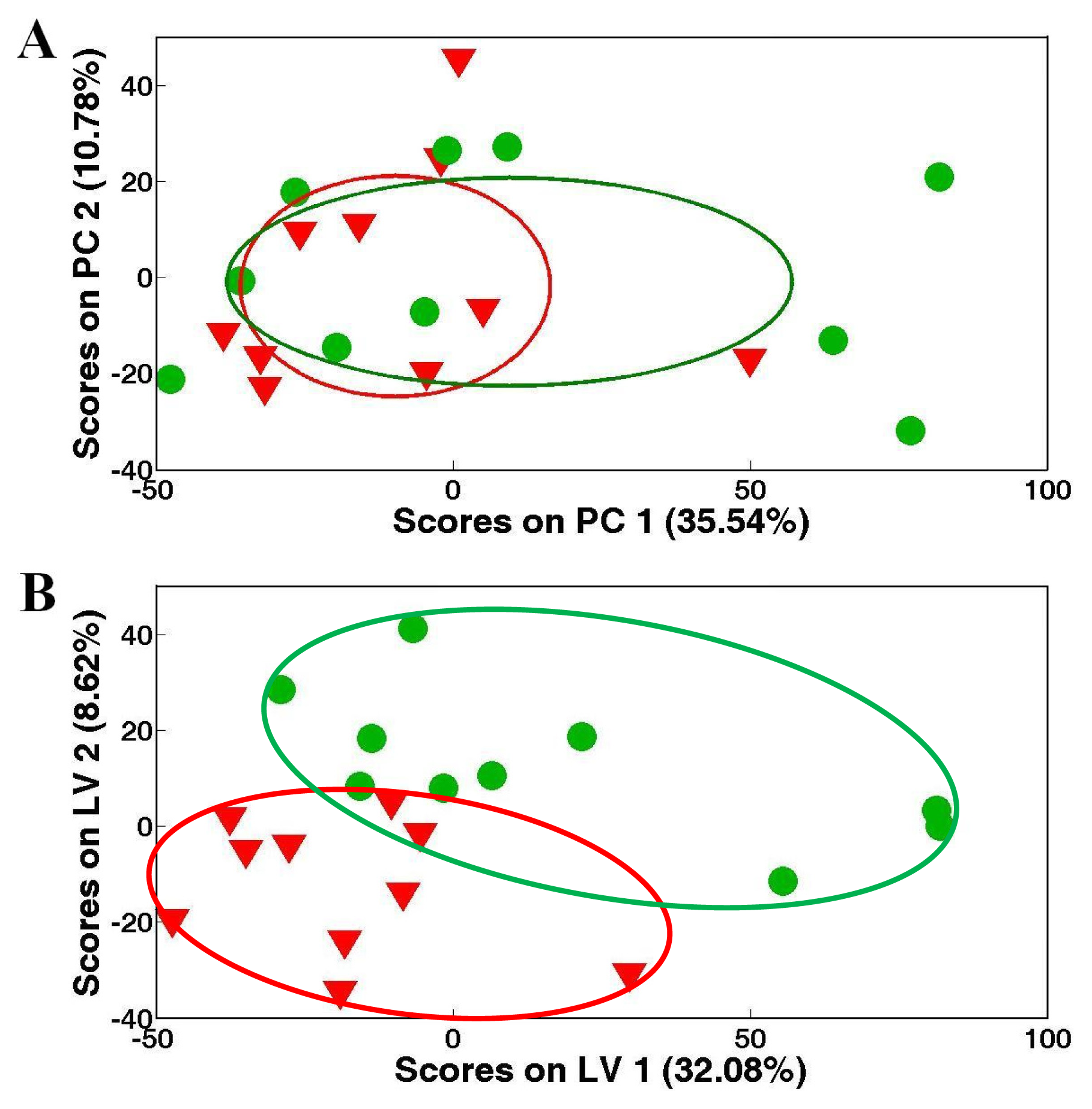
 ) White clams, and corresponding PC loadings plots, (B), (D) and (F) showing the metabolic differences between the control and heavy metal-exposed clam samples after exposure for 48 h. Ellipses represented mean ± standard deviation of PC scores for each group. Keys: (1) branched chain amino acids, (2) alanine, (3) unknown 1 (1.51 ppm), (4) 2-aminoadipate, (5) arginine, (6) glutamate, (7) succinate, (8) aspartate, (9) 4-aminobutyrate, (10) unknown 2 (3.14 ppm), (11) unknown 3 (3.72 ppm), (12) unknown 4 (4.12 ppm), (13) homarine, (14) β-glucose, (15) α-glucose, (16) lactate, (17) glutamine, (18) unknown 4 (3.16 ppm), (19) phosphocholine, (20) glycine, (21) unknown 5 (3.63 ppm), (22) taurine, (23) betaine and (24) ATP.
) White clams, and corresponding PC loadings plots, (B), (D) and (F) showing the metabolic differences between the control and heavy metal-exposed clam samples after exposure for 48 h. Ellipses represented mean ± standard deviation of PC scores for each group. Keys: (1) branched chain amino acids, (2) alanine, (3) unknown 1 (1.51 ppm), (4) 2-aminoadipate, (5) arginine, (6) glutamate, (7) succinate, (8) aspartate, (9) 4-aminobutyrate, (10) unknown 2 (3.14 ppm), (11) unknown 3 (3.72 ppm), (12) unknown 4 (4.12 ppm), (13) homarine, (14) β-glucose, (15) α-glucose, (16) lactate, (17) glutamine, (18) unknown 4 (3.16 ppm), (19) phosphocholine, (20) glycine, (21) unknown 5 (3.63 ppm), (22) taurine, (23) betaine and (24) ATP.
 ) and Cd (A), Zn (C) and mixed Cd and Zn (E) exposed (
) and Cd (A), Zn (C) and mixed Cd and Zn (E) exposed (
 ) White clams, and corresponding PC loadings plots, (B), (D) and (F) showing the metabolic differences between the control and heavy metal-exposed clam samples after exposure for 48 h. Ellipses represented mean ± standard deviation of PC scores for each group. Keys: (1) branched chain amino acids, (2) alanine, (3) unknown 1 (1.51 ppm), (4) 2-aminoadipate, (5) arginine, (6) glutamate, (7) succinate, (8) aspartate, (9) 4-aminobutyrate, (10) unknown 2 (3.14 ppm), (11) unknown 3 (3.72 ppm), (12) unknown 4 (4.12 ppm), (13) homarine, (14) β-glucose, (15) α-glucose, (16) lactate, (17) glutamine, (18) unknown 4 (3.16 ppm), (19) phosphocholine, (20) glycine, (21) unknown 5 (3.63 ppm), (22) taurine, (23) betaine and (24) ATP.
) White clams, and corresponding PC loadings plots, (B), (D) and (F) showing the metabolic differences between the control and heavy metal-exposed clam samples after exposure for 48 h. Ellipses represented mean ± standard deviation of PC scores for each group. Keys: (1) branched chain amino acids, (2) alanine, (3) unknown 1 (1.51 ppm), (4) 2-aminoadipate, (5) arginine, (6) glutamate, (7) succinate, (8) aspartate, (9) 4-aminobutyrate, (10) unknown 2 (3.14 ppm), (11) unknown 3 (3.72 ppm), (12) unknown 4 (4.12 ppm), (13) homarine, (14) β-glucose, (15) α-glucose, (16) lactate, (17) glutamine, (18) unknown 4 (3.16 ppm), (19) phosphocholine, (20) glycine, (21) unknown 5 (3.63 ppm), (22) taurine, (23) betaine and (24) ATP.
 ) and Cd (A), Zn (C) exposed (
) and Cd (A), Zn (C) exposed (
 ) Zebra clams, and corresponding PC loadings plots, (B), (D) showing the metabolic differences between the control and heavy metal-exposed clam samples after exposure for 48 h. Ellipses represented mean ± standard deviation of PC scores for each group. Keys: (1) branched chain amino acids, (2) lactate, (3) alanine, (4) unknown 1 (1.51 ppm), (5) glutamate, (6) glutamine, (7) acetoacetate, (8) phosphocholine, (7) aspartate, (9) unknown 2 (3.63 ppm), (10) unknown 3 (4.12 ppm), (11) homarine, (12) histidine, (13) unknown 4 (7.68 ppm), (14) succinate, (15) aspartate, (16) hypotaurine, (17) unknown 5 (3.72 ppm) and (18) ATP.
) Zebra clams, and corresponding PC loadings plots, (B), (D) showing the metabolic differences between the control and heavy metal-exposed clam samples after exposure for 48 h. Ellipses represented mean ± standard deviation of PC scores for each group. Keys: (1) branched chain amino acids, (2) lactate, (3) alanine, (4) unknown 1 (1.51 ppm), (5) glutamate, (6) glutamine, (7) acetoacetate, (8) phosphocholine, (7) aspartate, (9) unknown 2 (3.63 ppm), (10) unknown 3 (4.12 ppm), (11) homarine, (12) histidine, (13) unknown 4 (7.68 ppm), (14) succinate, (15) aspartate, (16) hypotaurine, (17) unknown 5 (3.72 ppm) and (18) ATP.
 ) and Cd (A), Zn (C) exposed (
) and Cd (A), Zn (C) exposed (
 ) Zebra clams, and corresponding PC loadings plots, (B), (D) showing the metabolic differences between the control and heavy metal-exposed clam samples after exposure for 48 h. Ellipses represented mean ± standard deviation of PC scores for each group. Keys: (1) branched chain amino acids, (2) lactate, (3) alanine, (4) unknown 1 (1.51 ppm), (5) glutamate, (6) glutamine, (7) acetoacetate, (8) phosphocholine, (7) aspartate, (9) unknown 2 (3.63 ppm), (10) unknown 3 (4.12 ppm), (11) homarine, (12) histidine, (13) unknown 4 (7.68 ppm), (14) succinate, (15) aspartate, (16) hypotaurine, (17) unknown 5 (3.72 ppm) and (18) ATP.
) Zebra clams, and corresponding PC loadings plots, (B), (D) showing the metabolic differences between the control and heavy metal-exposed clam samples after exposure for 48 h. Ellipses represented mean ± standard deviation of PC scores for each group. Keys: (1) branched chain amino acids, (2) lactate, (3) alanine, (4) unknown 1 (1.51 ppm), (5) glutamate, (6) glutamine, (7) acetoacetate, (8) phosphocholine, (7) aspartate, (9) unknown 2 (3.63 ppm), (10) unknown 3 (4.12 ppm), (11) homarine, (12) histidine, (13) unknown 4 (7.68 ppm), (14) succinate, (15) aspartate, (16) hypotaurine, (17) unknown 5 (3.72 ppm) and (18) ATP.
© 2011 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wu, H.; Liu, X.; Zhao, J.; Yu, J. NMR-Based Metabolomic Investigations on the Differential Responses in Adductor Muscles from Two Pedigrees of Manila Clam Ruditapes philippinarum to Cadmium and Zinc. Mar. Drugs 2011, 9, 1566-1579. https://doi.org/10.3390/md9091566
Wu H, Liu X, Zhao J, Yu J. NMR-Based Metabolomic Investigations on the Differential Responses in Adductor Muscles from Two Pedigrees of Manila Clam Ruditapes philippinarum to Cadmium and Zinc. Marine Drugs. 2011; 9(9):1566-1579. https://doi.org/10.3390/md9091566
Chicago/Turabian StyleWu, Huifeng, Xiaoli Liu, Jianmin Zhao, and Junbao Yu. 2011. "NMR-Based Metabolomic Investigations on the Differential Responses in Adductor Muscles from Two Pedigrees of Manila Clam Ruditapes philippinarum to Cadmium and Zinc" Marine Drugs 9, no. 9: 1566-1579. https://doi.org/10.3390/md9091566



