Biofilm Formation and Antimicrobial Susceptibility of Staphylococcus epidermidis Strains from a Hospital Environment
Abstract
:1. Introduction
2. Experimental Section
2.1. Analysis of Biofilm Production by the Congo Red Agar (CRA).
2.2. Microtiter Plate Assay (TCP)
2.3. Detection of icaADBC Genes S. epidermidis Strains
2.4. Antimicrobial Susceptibility Testing
2.5. Statistical Analysis
3. Results and Discussion
| Strain (No. of Strains) | a FOX | a E | a DA | a T | a C | a CIP | a CN | a RIF | a LNZ | a SXT |
|---|---|---|---|---|---|---|---|---|---|---|
| S. epidermidis (32) | 25/78.1 | 18/56.3 | 26/81.3 | 25/78.1 | 32/100 | 32/100 | 31/96.9 | 32/100 | 31/96.9 | 23/71.9 |
| S. haemolyticus (31) | 26/83.9 | 14/45.2 | 19/61.3 | 26/83.9 | 25/80.6 | 27/87.1 | 28/90.3 | 31/100 | 31/100 | 18/58.1 |
| S. capitis subsp. capitis (21) | 17/81 | 9/42.9 | 15/71.4 | 19/90.5 | 21/100 | 10/47.6 | 10/41.6 | 19/90.5 | 21/100 | 17/81 |
| S. hominis (11) | 10/90.9 | 4/36.4 | 9/81.8 | 6/54.5 | 9/81.8 | 11/100 | 11/100 | 11/100 | 11/100 | 8/72.7 |
| S. cohnii subsp. cohnii (9) | 7/77.8 | 5/55.6 | 5/55.6 | 5/55.6 | 9/100 | 9/100 | 9/100 | 9/100 | 9/100 | 5/55.6 |
| S. saprophyticus (5) | 5/100 | 1/20 | 4/80 | 5/100 | 5/100 | 5/100 | 5/100 | 5/100 | 5/100 | 3/60 |
| S. kloosii (4) | 4/100 | 2/50 | 3/75 | 1/25 | 3/75 | 4/100 | 4/100 | 4/100 | 4/100 | 4/100 |
| S. warneri (4) | 3/75 | 1/25 | 1/25 | 3/75 | 3/75 | 4/100 | 4/100 | 4/100 | 4/100 | 3/75 |
| S. cohnii subsp. urealyticum (2) | 2/100 | 0/0 | 1/50 | 1/50 | 2/100 | 2/100 | 2/100 | 2/100 | 2/100 | 1/50 |
| S. lugdunensis (1) | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 |
| S. hyicus (1) | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 | 0/0 |
| S. chromogenes (1) | 1/100 | 0/0 | 1/100 | 0/0 | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 | 1/100 |
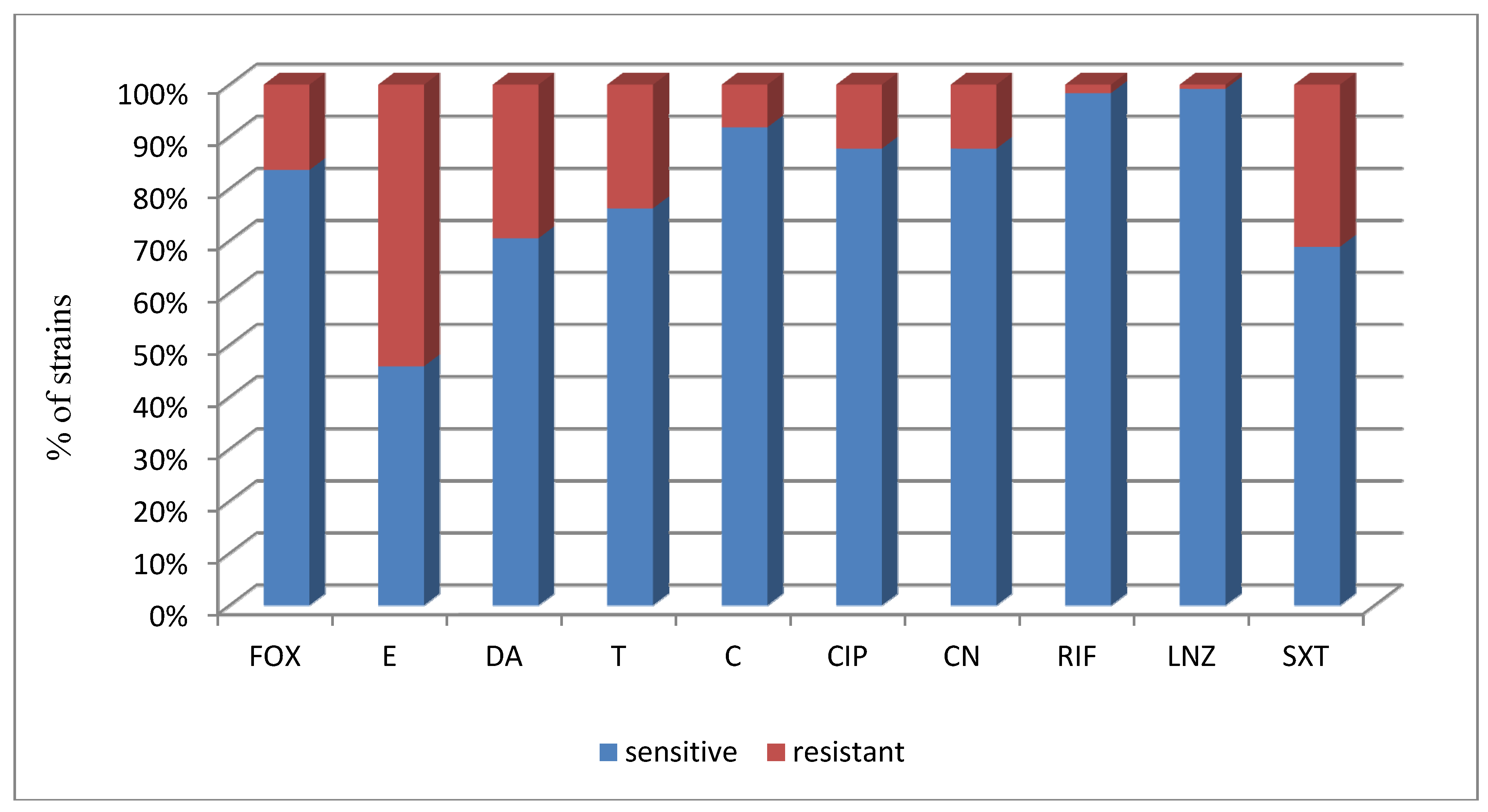
| Total (122)/% | 10/83.6 | 56/45.9 | 86/70.5 | 93/76.2 | 112/91.8 | 107/87.7 | 107/87.7 | 120/98.4 | 121/99.2 | 84/68.9 |
| Strain (No. of strains) | No (%) of Positive Strains, CRA Method | No (%) of Positive Strains, TCP Method OD > 0.17,—Strong Biofilm Production | No (%) of Positive Strains, TCP Method 0.11 > OD >0.16—Weak Biofilm Production |
|---|---|---|---|
| S. epidermidis (32) | 0 | 9 (28.1) | 3 (9.4) |
| S. haemolyticus (31) | 1 (3.2) | 1 (3.2) | 3 (3.2) |
| S. capitis subsp. capitis (21) | 3 (14.3) | 0 | 1 (4.8) |
| S. hominis (11) | 1 (9.1) | 0 | 2 (18.2) |
| S. cohnii subsp. cohnii (9) | 2 (22.2) | 0 | 0 |
| S. saprophyticus (5) | 0 | 0 | 0 |
| S. kloosii (4) | 1 (25) | 1 (25) | 1 (25) |
| S. warneri (4) | 0 | 0 | 1 (25) |
| S. cohnii subsp. urealyticum (2) | 0 | 1 (50) | 1 (50) |
| S. lugdunensis (1) | 0 | 0 | 0 |
| S. hyicus (1) | 1 (100) | 0 | 0 |
| S. chromogenes (1) | 0 | 0 | 1 (100) |
| Total: (122) | 9 (7.4) | 12 (9.8) | 13 (10.7) |
| a FOX | a E | a DA | a T | a C | a CIP | a CN | a RIF | a LNZ | a SXT | |
|---|---|---|---|---|---|---|---|---|---|---|
| S. epidermidis (32) | ||||||||||
| No. of susceptible strains | 25 | 18 | 26 | 25 | 32 | 32 | 31 | 32 | 31 | 23 |
| % of susceptible strains | 78.1% | 56.3% | 81.3% | 78.1% | 100% | 100% | 96.9% | 100% | 96.9% | 71.9% |
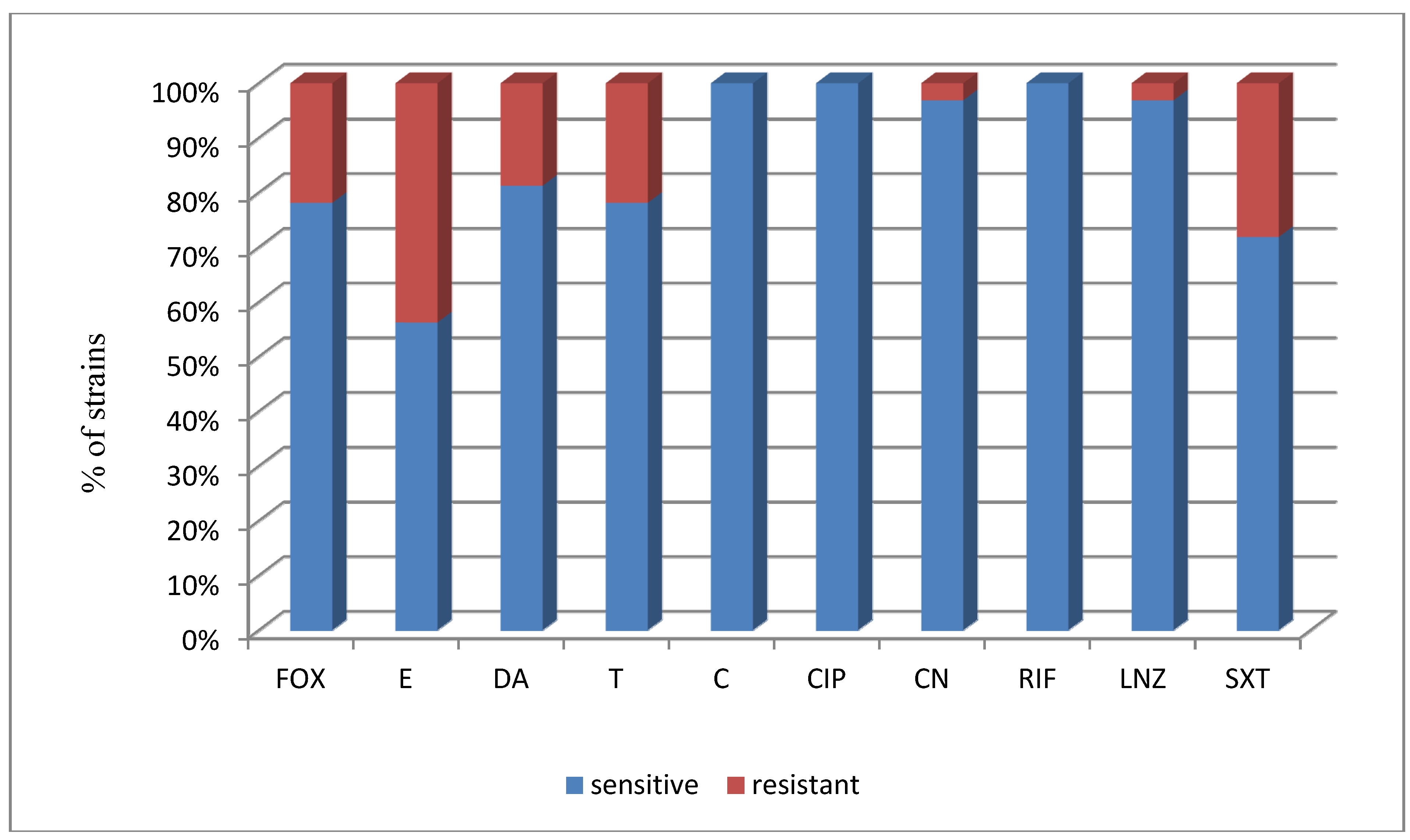
| biofilm strains (12) | ||||||||||
| No. of susceptible strains | 9 | 6 | 9 | 9 | 12 | 12 | 11 | 12 | 11 | 10 |
| % of susceptible strains | 75% | 50% | 75% | 75% | 100% | 100% | 91.7% | 100% | 91.7% | 83.3% |
| non biofilm strains (20) | ||||||||||
| No. of susceptible strains | 16 | 12 | 17 | 16 | 20 | 20 | 20 | 20 | 20 | 13 |
| % of susceptible strains | 80% | 60% | 85% | 80% | 100% | 100% | 100% | 100% | 100% | 65% |
| b P | 1.000 | 0.718 | 0.647 | 0.535 | c NA | NA | 0.375 | NA | 0.375 | 0.205 |
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- European Centre for Disease Prevention and Control (ECDC). Annual Epidemiological Report on Communicable Diseases in Europe; ECDC: Stockholm, Sweden, 2007. [Google Scholar]
- Lis, D.O.; Pacha, J.Z.; Idzik, D. Methicillin resistance of airbiorne coagulase-negative staphylococci in homes of persons having contact with a hospital environment. Am. J. Infect. Control 2009, 37, 177–182. [Google Scholar] [CrossRef]
- Bryers, J.D. Medical biofilms. Biotechnol. Bioeng. 2008, 100, 1–18. [Google Scholar] [CrossRef]
- Kampf, G.; Löffler, H.; Gastmeier, P. Hand hygiene for the prevention of nosocomial infections. Dtsch. Ärztebl. Int. 2009, 106, 649–655. [Google Scholar]
- Høiby, N.; Bjarnsholta, T.; Givskovb, M.; Molinc, S.; Ciofub, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef]
- Nieshimura, S.; Tsurumoto, T.; Yonekura, A.; Adachi, K.; Shindo, H. Antimicrobial susceptibility of Staphylococcus aureus and Staphylococcus epidermidis biofilms isolated from infected total hip arthroplasty case. J. Orthoped. Sci. 2006, 11, 46–50. [Google Scholar] [CrossRef]
- Presterl, E.; Suchomel, M.; Eder, M.; Reichmann, S.; Lassnigg, A.; Graninger, W.; Rotter, M. Effects of alcohols, povidone-iodine and hydrogen peroxide on biofilms of Staphylococcus epidermidis. J. Antimicrob. Chemother. 2007, 60, 417–420. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Izano, E.; Amarante, M.; Kher, W.; Kaplan, J. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 2008, 74, 470–476. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis—the “accidental” pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef]
- Mack, D.; Fischer, W.; Krokotsch, A.; Leopold, K.; Hartmann, R.; Egge, H.; Laufs, R. The intercellular adhesin involved in biofilm accumulation Staphylococcus epidermidis linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 1996, 178, 175–183. [Google Scholar]
- Darby, C.; Hsu, J.W.; Ghori, N.; Falkow, S. Caenorhabditis. elegans: Plague bacteria biofilm blocks food intake. Nature 2002, 16, 243–244. [Google Scholar]
- Kaplan, J.B.; Velliyagounder, K.; Ragunath, Ch.; Rode, H.; Mack, D.; Knobloch, J.K.; Ramasbbu, N. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus. actinomycetemcomitans and Actinobacillus. pleuropneumoniae biofilms. J. Bacteriol. 2004, 186, 8213–8220. [Google Scholar] [CrossRef]
- Wang, X.; Preston, J.F.I.; Romeo, T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 2004, 186, 2724–2734. [Google Scholar] [CrossRef]
- Vuong, C.; Kocianova, S.; Voyich, J.M.; Yao, Y.; Fishcer, E.R.; DeLeo, F.R.; Otto, M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 2004, 279, 54881–54886. [Google Scholar]
- Gerke, C.; Kraft, A.; Sussmuth, R.; Schweitzer, O.; Götz, F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesion. J. Biol. Chem. 1998, 273, 18586–18593. [Google Scholar]
- Rupp, M.E.; Fey, P.D.; Heilmann, C.; Götz, F. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J. Infect. Dis. 2001, 183, 1038–1042. [Google Scholar] [CrossRef]
- Fluckiger, U.; Ulrich, M.; Steiuhuber, A.; Döring, G.; Mack, D.; Landmann, R.; Goerke, Ch.; Wolz, Ch. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect. Immun. 2005, 73, 1811–1819. [Google Scholar] [CrossRef]
- Costa, F.S.; Miceli, M.H.; Anaissie, E.J. Mucosa or skin as source of coagulase-negative staphylococcal bacteraemia. Lancet Infect. Dis. 2004, 4, 278–286. [Google Scholar] [CrossRef]
- Klingenberg, C.; Aarag, E.; Ronnestad, A.; Sollid, J.E.; Abrahamsen, M.D.; Kjeldsen, G.; Flaegstad, T. Coagulase-negative staphylococcal sepsis in neonates—Association between antibiotic resistance, biofilm formation and the host inflammatory response. Pediatr. Infect. Dis. J. 2005, 24, 817–822. [Google Scholar] [CrossRef]
- Oliveira, A.; Cunha, M.L.R.S. Comparison of methods for the detection of biofilm production in coagulase-negative staphylococci. BMC Res. Notes 2010, 3. [Google Scholar] [CrossRef]
- O’Gara, J.; Humphreys, H. Staphylococcus epidermidis biofilms: Importance and implications. J. Med. Microbiol. 2001, 50, 582–587. [Google Scholar]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Brit. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef]
- Hart, J.B.; French, M.L.V.; Eitzen, H.E.; Ritter, M.A. Rodac plate-holding device for sampling surfaces during surgery. Appl. Microbiol. 1973, 26, 417–418. [Google Scholar]
- Lemmen, S.W.; Häfner, H.; Zolldann, D.; Amedick, G.; Lutticken, R. Comparison of two sampling methods for the detection of Gram-positive and Gram-negative bacteria in the environment: Moistened swabs versus Rodac plates. Int. J. Hyg. Environ. Health 2001, 203, 245–248. [Google Scholar] [CrossRef]
- Freeman, D.J.; Falkiner, F.R.; Keane, C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989, 42, 872–874. [Google Scholar] [CrossRef]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microb. 1985, 22, 996–1006. [Google Scholar]
- Ziebuhr, W.; Krimmer, V.; Rachid, S.; Lößner, I.; Götz, F.; Hacker, J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: Evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 1999, 32, 345–356. [Google Scholar] [CrossRef]
- De Silva, G.D.I.; Kantzanou, M.; Justice, A.; Massey, R.C.; Wilkinson, A.R.; Day, N.P.J.; Peacock, S.J. The ica operon and biofilm production in coagulase-negative staphylococci associated with carriage and disease in a neonatal intensive care unit. J. Clin. Microbiol. 2002, 40, 382–388. [Google Scholar] [CrossRef]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). Terminology relating to methods for the determination of susceptibility of bacteria to antimicrobial agents. EUCAST definitive document E. Def 1.2. Clin. Microbiol. Infect. 2000, 6, 503–508. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 207–228. [Google Scholar]
- Mathur, T.; Singhal, S.; Khan, S.; Upadhyay, D.J.; Fatma, T.; Rattan, A. Detection of biofilm formation among the clinical isolates of staphylococci: An evaluation of three different screening methods. Ind. J. Med. Microbiol. 2006, 24, 25–29. [Google Scholar] [CrossRef]
- Wojtyczka, R.D.; Krakowian, D.; Marek, Ł.; Skiba, D.; Kudelski, A.; Jasik, K.; Pacha, J. Analysis of the polymorphism of Staphylococcus strains isolated from a hospital environment. Afr. J. Microbiol. Res. 2011, 5, 4997–5003. [Google Scholar]
- Fredheim, E.G.A.; Klingenberg, C.; Rohde, H.; Frankenberger, S.; Gaustad, P.; Flægstad, T.; Solid, J.E. Biofilm Formation by Staphylococcus haemolyticus. J. Clin. Microbiol. 2009, 47, 1172–1180. [Google Scholar]
- Arciola, C.R.; Baldassarri, L.; Montanaro, L. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 2001, 39, 2151–2156. [Google Scholar] [CrossRef]
- Růžička, F.; Holá, V.; Vota, M.; Tejkalová, R.; Horvát, R.; Heroldová, M.; Woznicowá, V. Biofilm detection and the clinical significance of Staphylococcus epidermidis isolates. Folia Microbiol. 2004, 49, 596–600. [Google Scholar] [CrossRef]
- Qin, Z.; Yang, X.; Yang, L.; Jiang, J.; Ou, Y.; Molin, S.; Qu, D. Formation and properties of in vitro biofilms of ica-negative Staphylococcus epidermidis clinical isolates. J. Med. Microbiol. 2007, 1, 83–93. [Google Scholar]
- Eftekhar, F.; Mirmohamadi, Z. Evaluation of biofilm production by Staphylococcus epidermidis isolates from nosocomial infections and skin of healthy volunteers. Int. J. Med. Med. Sci. 2009, 1, 438–441. [Google Scholar]
- Kochman, M. Susceptibility of the bacteria isolated from samples of clinical material in Poland in 1998 to selected chemotherapeutics and antibiotics. The analysis of the questionnaire findings. I. Susceptibility of staphylococci. Przegl. Epidemiol. 2005, 59, 679–694. [Google Scholar]
- Michnowska-Swincow, E.; Szychlińska, I. Drug resistance of methicillin resistant coagulase-negative staphylococci (MRCNS) isolated from hospital environment. Diag. Labor. 2001, 37, 437–444. [Google Scholar]
- Piette, A.; Verschraegen, G. Role of coagulase-negative staphylococci in human disease. Vet. Microbiol. 2009, 134, 45–54. [Google Scholar] [CrossRef]
- Tunger, O.; Ozbakkaloglu, B.; Aksoy, H. Trends in antimicrobial resistant staphylococci in an university hospital over a 6-year period. Int. J. Antimicrob. Agents 2001, 18, 93–96. [Google Scholar]
- Olivares, M.J.; Orozco, R.H.; Garrido, S.R.; Rodríguez-Vidigal, F.F.; Tomé, A.V.; Marcos, M.R. Activity of vancomycin, ciprofloxacin, daptomycin and linezolid against coagulasenegative staphylococci bacteremia. Rev. Esp. Quimioter. 2011, 24, 74–78. [Google Scholar]
- Sarathbabu, R.; Rajkumari, N.; RamaNi, V. Characterization of Coagulase negative Staphylococci isolated from urine, pus, sputum and blood samples. Int. J. Pharma. Sci. Inv. 2013, 2, 37–46. [Google Scholar]
- Zheng, Z.; Stewart, P.S. Penetration of Rifampin through Staphylococcus epidermidis Biofilms. Antimicrob. Agents Chemother. 2002, 46, 900–903. [Google Scholar] [CrossRef]
- Leite, B.; Gomes, F.; Teixeira, P.; Souza, C.; Pizzolitto, E.; Oliveira, R. In vitro activity of daptomycin, linezolid and rifampicin on Staphylococcus epidermidis biofilms. Curr. Microbiol. 2011, 63, 313–317. [Google Scholar] [CrossRef] [Green Version]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wojtyczka, R.D.; Orlewska, K.; Kępa, M.; Idzik, D.; Dziedzic, A.; Mularz, T.; Krawczyk, M.; Miklasińska, M.; Wąsik, T.J. Biofilm Formation and Antimicrobial Susceptibility of Staphylococcus epidermidis Strains from a Hospital Environment. Int. J. Environ. Res. Public Health 2014, 11, 4619-4633. https://doi.org/10.3390/ijerph110504619
Wojtyczka RD, Orlewska K, Kępa M, Idzik D, Dziedzic A, Mularz T, Krawczyk M, Miklasińska M, Wąsik TJ. Biofilm Formation and Antimicrobial Susceptibility of Staphylococcus epidermidis Strains from a Hospital Environment. International Journal of Environmental Research and Public Health. 2014; 11(5):4619-4633. https://doi.org/10.3390/ijerph110504619
Chicago/Turabian StyleWojtyczka, Robert D., Kamila Orlewska, Małgorzata Kępa, Danuta Idzik, Arkadiusz Dziedzic, Tomasz Mularz, Michał Krawczyk, Maria Miklasińska, and Tomasz J. Wąsik. 2014. "Biofilm Formation and Antimicrobial Susceptibility of Staphylococcus epidermidis Strains from a Hospital Environment" International Journal of Environmental Research and Public Health 11, no. 5: 4619-4633. https://doi.org/10.3390/ijerph110504619






