Lipoprotein Lipase (LPL) Polymorphism and the Risk of Coronary Artery Disease: A Meta-Analysis
Abstract
:1. Introduction
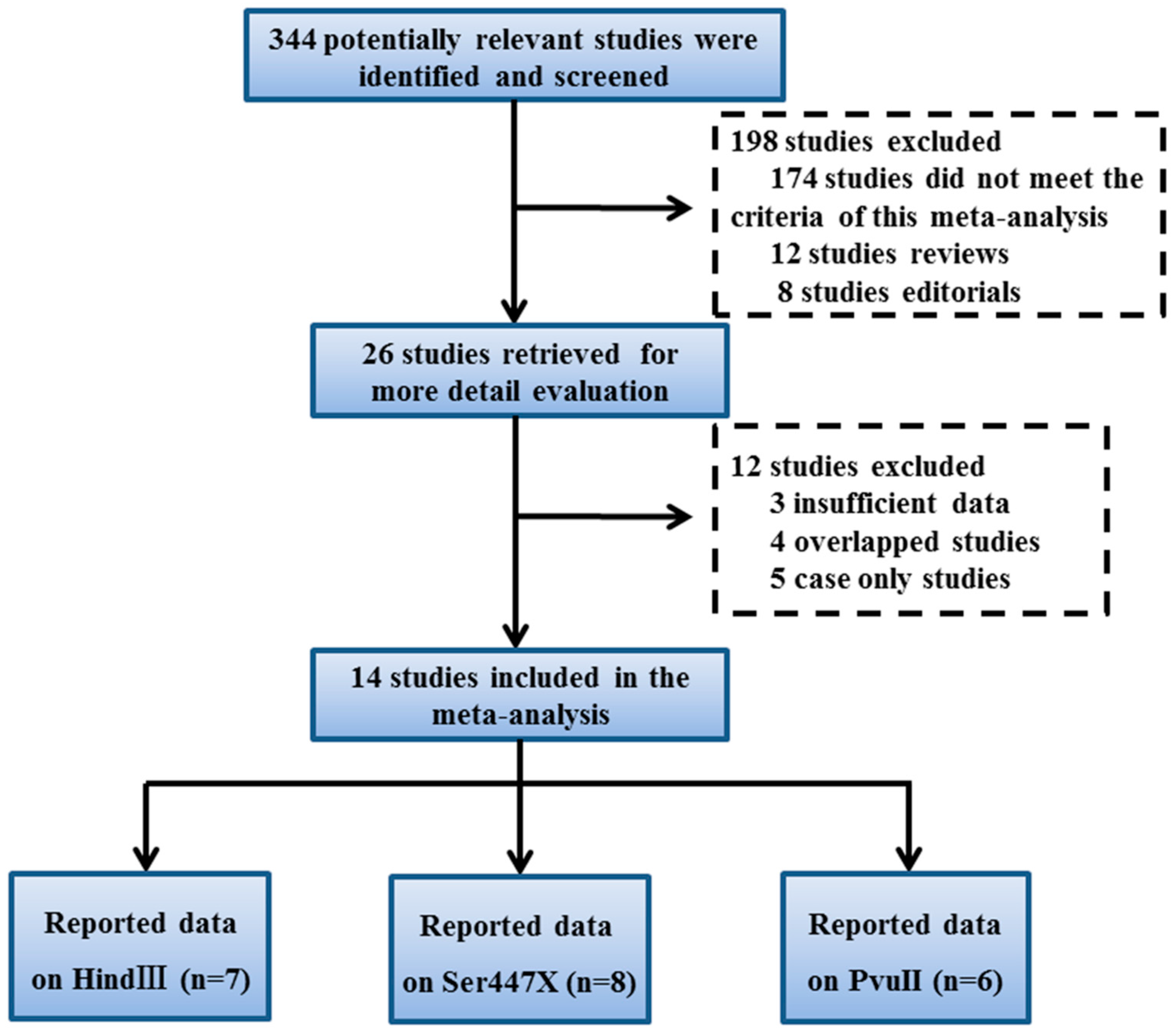
2. Materials and Methods
2.1. Literature Sources and Search Strategies
2.2. Inclusion and Exclusion Criteria
2.3. Data Extraction
2.4. Statistical Analyses
3. Results
3.1. Study Characteristics
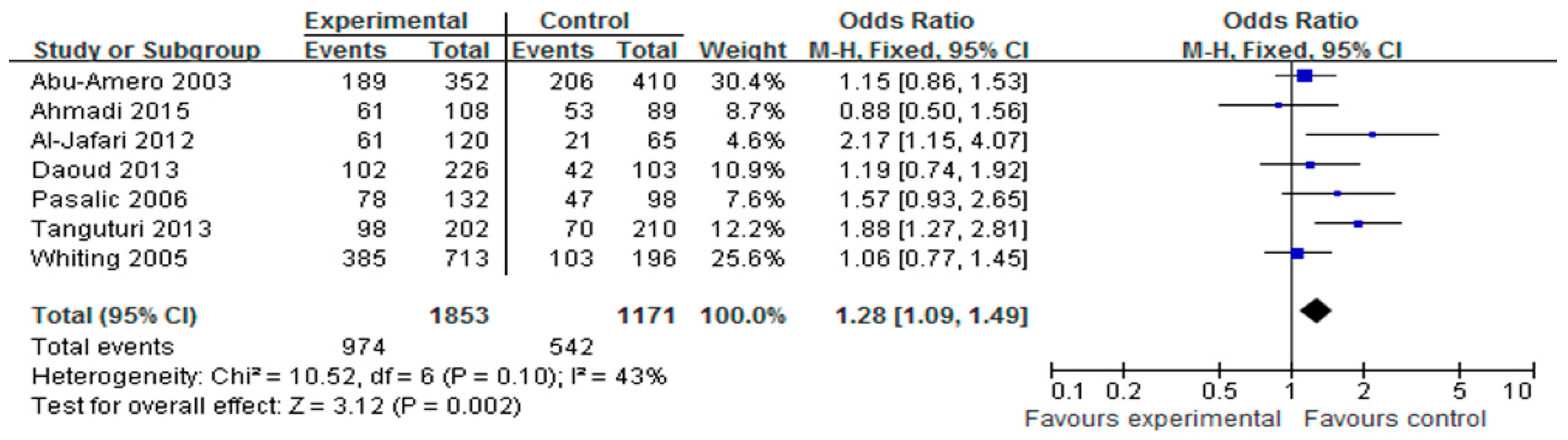
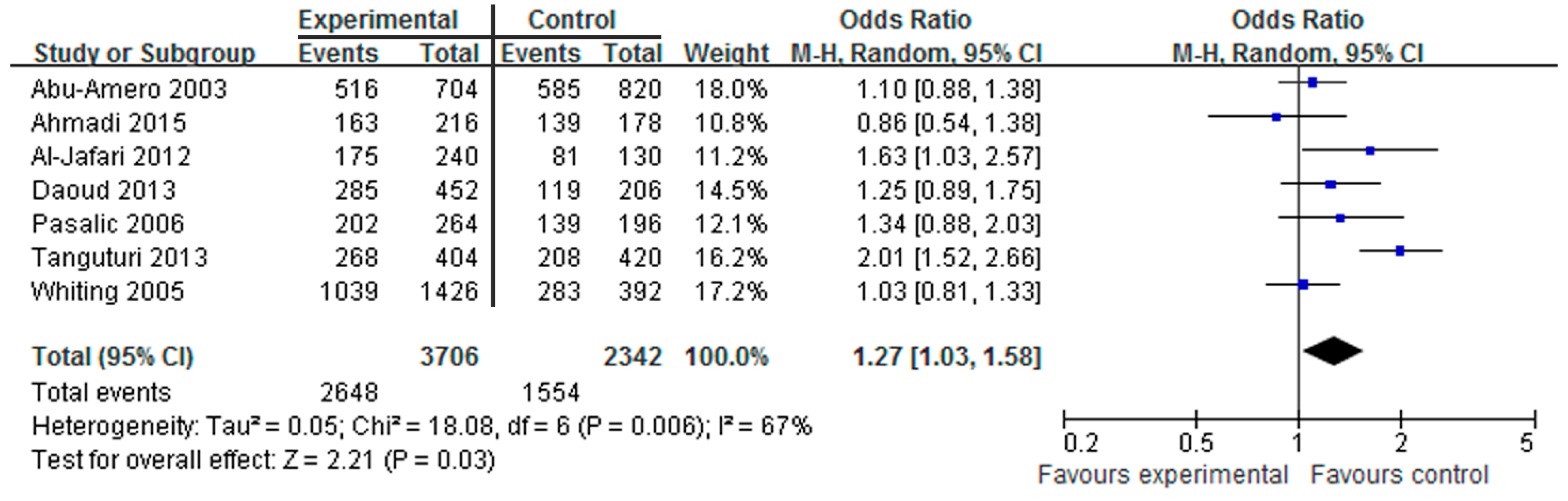
3.2. Association of the HindIII Polymorphism with CAD
3.3. Association of the Ser447X Polymorphism with CAD
3.4. Association of the PvuII Polymorphism with CAD
3.5. Sensitivity Analyses and Publication Bias
4. Discussion
5. Conclusions
Author Contributions
Conflicts of Interests
References
- Ardeshiri, M.; Faritus, Z.; Ojaghi-Haghighi, Z.; Bakhshandeh, H.; Kargar, F.; Aghili, R. Impact of metabolic syndrome on mortality and morbidity after coronary artery bypass grafting surgery. Res. Cardiovasc. Med. 2014, 3, e20270. [Google Scholar] [CrossRef] [PubMed]
- Teo, K.K.; Cohen, E.; Buller, C.; Hassan, A.; Carere, R.; Cox, J.L.; Ly, H.; Fedak, P.W.; Chan, K.; Legare, J.F.; et al. Canadian Cardiovascular Society/Canadian Association of Interventional Cardiology/Canadian Society of Cardiac Surgery position statement on revascularization-multivessel coronary artery disease. Can. J. Cardiol. 2014, 30, 1482–1491. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.L.; Hsu, L.A.; Hsu, K.H.; Ko, Y.H.; Lee, Y.S. The interactive effects of hepatic lipase gene promoter polymorphisms with sex and obesity on high-density-lipoprotein cholesterol levels in Taiwanese-Chinese. Atherosclerosis 2004, 172, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Clee, S.M.; Kastelein, J.J.; van Dam, M.; Marcil, M.; Roomp, K.; Zwarts, K.Y.; Collins, J.A.; Roelants, R.; Tamasawa, N.; Stulc, T.; et al. Age and residual cholesterol efflux affect HDL cholesterol levels and coronary artery disease in ABCA1 heterozygotes. J. Clin. Investig. 2000, 106, 1263–1270. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.Y.; Li, S.; Wang, M.; Qu, X.; Hu, G.; Xu, Z.; Chen, M.; He, G.W.; Wu, H. Association of monocyte chemoattractant protein-1 (MCP-1)-2518A>G polymorphism with susceptibility to coronary artery disease: A meta-analysis. Ann. Hum. Genet. 2015, 79, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Xie, X.; Ma, Y.T.; Zheng, Y.Y.; Yang, Y.N.; Li, X.M.; Fu, Z.Y.; Liu, F.; Chen, B.D. Association of COX-2-765G>C genetic polymorphism with coronary artery disease: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 7412–7418. [Google Scholar] [PubMed]
- Kusunoki, M.; Tsutsumi, K.; Sato, D.; Nakamura, A.; Habu, S.; Mori, Y.; Morishita, M.; Yonemoto, T.; Miyata, T.; Nakaya, Y.; et al. Activation of lipoprotein lipase increases serum high density lipoprotein 2 cholesterol and enlarges high density lipoprotein 2 particles in rats. Eur. J. Pharmacol. 2011, 668, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tong, J.; Li, S.; Zhang, R.; Chen, L.; Wang, Y.; Zheng, M.; Wang, M.; Liu, G.; Dai, Y.; et al. Over-expression of human lipoprotein lipase in mouse mammary glands leads to reduction of milk triglyceride and delayed growth of suckling pups. PLoS ONE 2011, 6, e20895. [Google Scholar] [CrossRef] [PubMed]
- Voss, C.V.; Davies, B.S.; Tat, S.; Gin, P.; Fong, L.G.; Pelletier, C.; Mottler, C.D.; Bensadoun, A.; Beigneux, A.P.; Young, S.G. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc. Natl. Acad. Sci. USA 2011, 108, 7980–7984. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Rimm, E.B.; Rader, D.; Schmidt, E.B.; Sorensen, T.I.; Vogel, U.; Overvad, K.; Mukamal, K.J. S447X variant of the lipoprotein lipase gene, lipids, and risk of coronary heart disease in 3 prospective cohort studies. Am. Heart J. 2009, 157, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Wion, K.L.; Kirchgessner, T.G.; Lusis, A.J.; Schotz, M.C.; Lawn, R.M. Human lipoprotein lipase complementary DNA sequence. Science 1987, 235, 1638–1641. [Google Scholar] [CrossRef] [PubMed]
- Sayad, A.; Noruzinia, M.; Zamani, M.; Harirchian, M.H.; Kazemnejad, A. Lipoprotein Lipase HindIII Intronic Polymorphism in a Subset of Iranian Patients with Late-Onset Alzheimer’s Disease. Cell J. 2012, 14, 67–72. [Google Scholar] [PubMed]
- Tanguturi, P.R.; Pullareddy, B.; Rama Krishna, B.S.; Murthy, D.K. Lipoprotein lipase gene HindIII polymorphism and risk of myocardial infarction in South Indian population. Indian Heart J. 2013, 65, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Abu-Amero, K.K.; Wyngaard, C.A.; Al-Boudari, O.M.; Kambouris, M.; Dzimiri, N. Lack of association of lipoprotein lipase gene polymorphisms with coronary artery disease in the Saudi Arab population. Arch. Pathol. Lab. Med. 2003, 127, 597–600. [Google Scholar] [PubMed]
- Al-Jafari, A.A.; Daoud, M.S.; Mobeirek, A.F.; Al Anazi, M.S. DNA polymorphisms of the lipoprotein lipase gene and their association with coronary artery disease in the Saudi population. Int. J. Mol. Sci. 2012, 13, 7559–7574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgiev, A.; Panov, S.; Sadikario, S. Association of PvuII polymorphism in the lipoprotein lipase gene with the coronary artery disease in Macedonian population. Prilozi 2008, 29, 213–225. [Google Scholar] [PubMed]
- Duman, B.S.; Turkoglu, C.; Akpinar, B.; Guden, M.; Vertii, A.; Dak, E.; Cagatay, P.; Gunay, D.; Buyukdevrim, A.S. Lipoprotein lipase gene polymorphism and lipid profile in coronary artery disease. Arch. Pathol. Lab. Med. 2004, 128, 869–874. [Google Scholar] [PubMed]
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials. Control. Clin. Trials 1986, 7, 177–188. [Google Scholar] [CrossRef]
- Mantel, N.; Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959, 22, 719–748. [Google Scholar] [PubMed]
- Egger, M.; Davey Smith, G.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Begg, C.B.; Mazumdar, M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994, 50, 1088–1101. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, Z.; Senemar, S.; Toosi, S.; Radmanesh, S. The association of lipoprotein lipase genes, HindIII and S447X polymorphisms with coronary artery disease in Shiraz city. J. Cardiovasc. Thorac. Res. 2015, 7, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Almeida, K.A.; Strunz, C.M.; Maranhao, R.C.; Mansur, A.P. The S447X polymorphism of lipoprotein lipase: Effect on the incidence of premature coronary disease and on plasma lipids. Arq. Bras. Cardiol. 2007, 88, 297–303. [Google Scholar] [PubMed]
- Aydogan, H.Y.; Isbir, S.; Kurnaz, O.; Gormus, U.; Isbir, T. Associations of lipoprotein lipase S447X and apolipoprotein E genotypes with low-density lipoprotein subfractions in Turkish patients with coronary artery disease. In Vivo 2009, 23, 155–161. [Google Scholar] [PubMed]
- Daoud, M.S.; Ataya, F.S.; Fouad, D.; Alhazzani, A.; Shehata, A.I.; Al-Jafari, A.A. Associations of three lipoprotein lipase gene polymorphisms, lipid profiles and coronary artery disease. Biomed. Rep. 2013, 1, 573–582. [Google Scholar] [PubMed]
- Ferencak, G.; Pasalic, D.; Grskovic, B.; Cheng, S.; Fijal, B.; Sesto, M.; Skodlar, J.; Rukavina, A.S. Lipoprotein lipase gene polymorphisms in Croatian patients with coronary artery disease. Clin. Chem. Lab. Med. 2003, 41, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Isbir, T.; Yilmaz, H.; Agachan, B.; Karaali, Z.E. Cholesterol ester transfer protein, apolipoprotein E and lipoprotein lipase genotypes in patients with coronary artery disease in the Turkish population. Clin. Genet. 2003, 64, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Pasalic, D.; Ferencak, G.; Grskovic, B.; Sesto, M.; Stavljenic-Rukavina, A. Association of two genetic variations of lipoprotein lipase, S447X and Hind III, with coronary artery disease and hypertriglyceridemia. Coll. Antropol. 2006, 30, 549–554. [Google Scholar] [PubMed]
- Sawano, M.; Watanabe, Y.; Ohmura, H.; Shimada, K.; Daida, H.; Mokuno, H.; Yamaguchi, H. Potentially protective effects of the Ser447-Ter mutation of the lipoprotein lipase gene against the development of coronary artery disease in Japanese subjects via a beneficial lipid profile. Jpn. Circ. J. 2001, 65, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Whiting, B.M.; Anderson, J.L.; Muhlestein, J.B.; Horne, B.D.; Bair, T.L.; Pearson, R.R.; Carlquist, J.F.; Intermountain Heart Collaborative Study Group. Candidate gene susceptibility variants predict intermediate end points but not angiographic coronary artery disease. Am. Heart J. 2005, 150, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Corella, D.; Guillen, M.; Saiz, C.; Portoles, O.; Sabater, A.; Folch, J.; Ordovas, J.M. Associations of LPL and APOC3 gene polymorphisms on plasma lipids in a Mediterranean population: Interaction with tobacco smoking and the APOE locus. J. Lipid Res. 2002, 43, 416–427. [Google Scholar] [PubMed]
- Socquard, E.; Durlach, A.; Clavel, C.; Nazeyrollas, P.; Durlach, V. Association of HindIII and PvuII genetic polymorphisms of lipoprotein lipase with lipid metabolism and macrovascular events in type 2 diabetic patients. Diabetes Metab. 2006, 32, 262–269. [Google Scholar] [CrossRef]
- Nicklas, B.J.; Ferrell, R.E.; Rogus, E.M.; Berman, D.M.; Ryan, A.S.; Dennis, K.E.; Goldberg, A.P. Lipoprotein lipase gene variation is associated with adipose tissue lipoprotein lipase activity, and lipoprotein lipid and glucose concentrations in overweight postmenopausal women. Hum. Genet. 2000, 106, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Razzaghi, H.; Demirci, F.Y.; Kamboh, M.I. Functional significance of lipoprotein lipase HindIII polymorphism associated with the risk of coronary artery disease. Atherosclerosis 2008, 200, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Humphries, S.E.; Nicaud, V.; Margalef, J.; Tiret, L.; Talmud, P.J. Lipoprotein lipase gene variation is associated with a paternal history of premature coronary artery disease and fasting and postprandial plasma triglycerides: The European Atherosclerosis Research Study (EARS). Arterioscler. Thromb. Vasc. Biol. 1998, 18, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Ukkola, O.; Garenc, C.; Perusse, L.; Bergeron, J.; Despres, J.P.; Rao, D.C.; Bouchard, C. Genetic variation at the lipoprotein lipase locus and plasma lipoprotein and insulin levels in the Quebec Family Study. Atherosclerosis 2001, 158, 199–206. [Google Scholar] [CrossRef]
- Oka, K.; Tkalcevic, G.T.; Stocks, J.; Galton, D.J.; Brown, W.V. Nucleotide sequence of PvuII polymorphic site at the human lipoprotein lipase gene locus. Nucleic Acids Res. 1989, 17, 6752. [Google Scholar] [CrossRef] [PubMed]
- Murthy, V.; Julien, P.; Gagne, C. Molecular pathobiology of the human lipoprotein lipase gene. Pharmacol. Ther. 1996, 70, 101–135. [Google Scholar] [CrossRef]

| Study | Years | Ethnicity | No. of Case/Control | Matching Criteria | Control Source | Genotyping | HWE |
|---|---|---|---|---|---|---|---|
| HindⅢ Polymorphism | |||||||
| Abu-Amero [14] | 2003 | Saudi Arabian | 352/410 | Sex, Age | HB | PCR | Yes |
| Ahmadi [22] | 2015 | Iranian | 108/89 | Sex, Age | HB | PCR | Yes |
| Al-Jafari [15] | 2012 | Saudi Arabian | 120/65 | Sex, Age | HB | PCR | Yes |
| Daoud [25] | 2013 | Saudi Arabian | 226/103 | Sex, Age | HB | PCR | Yes |
| Pasalic [28] | 2006 | Croatian | 132/98 | Sex, Age | HB | PCR | Yes |
| Tanguturi [13] | 2013 | Indian | 202/210 | Sex, Age | HB | PCR | Yes |
| Whiting [30] | 2004 | American | 713/196 | Sex, Age | HB | PCR | Yes |
| PvuII Polymorphism | |||||||
| Abu-Amero [14] | 2003 | Saudi Arabian | 431/511 | Sex, Age | HB | PCR | Yes |
| Al-Jafari [15] | 2012 | Saudi Arabian | 120/65 | Sex, Age | HB | PCR | Yes |
| Daoud [25] | 2013 | Saudi Arabian | 226/103 | Sex, Age | HB | PCR | Yes |
| Duman [17] | 2004 | Turkish | 78/49 | Sex, Age | HB | PCR | Yes |
| Georgiev [16] | 2008 | Macedonian | 109/32 | Sex, Age | HB | PCR | Yes |
| Isbir [27] | 2003 | Turkish | 100/72 | Sex, Age | HB | PCR | Yes |
| Ser447X Polymorphism | |||||||
| Ahmadi [22] | 2015 | Iranian | 115/89 | Sex, Age | HB | PCR | Yes |
| Al-Jafari [15] | 2012 | Saudi Arabian | 120/65 | Sex, Age | HB | PCR | Yes |
| Almeida [23] | 2006 | Brazil | 313/150 | Sex, Age | HB | PCR | Yes |
| Aydogan [24] | 2009 | Turkey | 41/23 | Sex, Age | HB | PCR | Yes |
| Daoud [25] | 2013 | Saudi Arabian | 226/103 | Sex, Age | HB | PCR | Yes |
| Ferencak [26] | 2003 | Croatian | 479/200 | Sex, Age | HB | PCR | Yes |
| Pasalic [28] | 2006 | Croatian | 132/98 | Sex, Age | HB | PCR | Yes |
| Sawano [29] | 2001 | Japanese | 93/96 | Sex, Age | HB | PCR | Yes |
| Author | Year | Case | Control | Sample Size | HWE (P) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HindⅢ Polymorphism | H+H+ | H+H− | H−H− | H+ | H− | H+H+ | H+H− | H−H− | H+ | H− | |||
| Abu-Amero [14] | 2003 | 189 | 138 | 25 | 516 | 188 | 206 | 173 | 31 | 585 | 235 | 352/410 | Yes |
| Pasalic [28] | 2006 | 78 | 46 | 8 | 202 | 62 | 47 | 45 | 6 | 139 | 57 | 132/98 | Yes |
| Whiting [30] | 2005 | 385 | 269 | 59 | 1039 | 387 | 103 | 77 | 16 | 283 | 109 | 713/196 | Yes |
| Daoud [25] | 2013 | 102 | 81 | 43 | 285 | 167 | 42 | 35 | 26 | 119 | 87 | 226/103 | Yes |
| Al-Jafari [15] | 2012 | 61 | 53 | 6 | 175 | 65 | 29 | 23 | 13 | 81 | 49 | 120/65 | Yes |
| Tanguturi [13] | 2013 | 98 | 72 | 32 | 268 | 136 | 70 | 68 | 72 | 208 | 212 | 202/210 | Yes |
| Ahmadi [22] | 2015 | 61 | 41 | 6 | 163 | 53 | 53 | 33 | 3 | 139 | 39 | 108/89 | Yes |
| Ser447X Polymorphism | SS | SX | XX | S | X | SS | SX | XX | S | X | |||
| Ahmadi [22] | 2015 | 58 | 23 | 34 | 139 | 91 | 75 | 7 | 7 | 157 | 21 | 115/89 | Yes |
| Al-Jafari [15] | 2012 | 100 | 20 | 0 | 220 | 20 | 57 | 8 | 0 | 122 | 8 | 120/65 | Yes |
| Almeida [23] | 2007 | 257 | 47 | 9 | 561 | 65 | 115 | 34 | 1 | 264 | 36 | 313/150 | Yes |
| Aydogan [24] | 2009 | 27 | 10 | 4 | 64 | 18 | 17 | 2 | 4 | 36 | 10 | 41/23 | Yes |
| Daoud [25] | 2013 | 185 | 41 | 0 | 411 | 41 | 92 | 11 | 0 | 195 | 11 | 226/103 | Yes |
| Ferencak [26] | 2003 | 378 | 97 | 4 | 853 | 105 | 167 | 32 | 1 | 366 | 34 | 479/200 | Yes |
| Pasalic [28] | 2006 | 113 | 19 | 0 | 245 | 19 | 69 | 28 | 1 | 166 | 30 | 132/98 | Yes |
| Sawano [29] | 2001 | 82 | 10 | 1 | 174 | 12 | 71 | 23 | 2 | 145 | 27 | 93/96 | Yes |
| PvuII Polymorphism | P+P+ | P+P− | P−P− | P+ | P− | P+P+ | P+P− | P−P− | P+ | P− | |||
| Abu-Amero [14] | 2003 | 138 | 225 | 68 | 501 | 361 | 182 | 248 | 81 | 612 | 410 | 431/511 | Yes |
| Al-Jafari [15] | 2012 | 50 | 52 | 18 | 152 | 88 | 25 | 28 | 12 | 78 | 52 | 120/65 | Yes |
| Daoud [25] | 2013 | 89 | 102 | 35 | 280 | 172 | 46 | 44 | 13 | 136 | 70 | 226/103 | Yes |
| Duman [17] | 2004 | 25 | 39 | 14 | 89 | 67 | 14 | 16 | 19 | 44 | 54 | 78/49 | Yes |
| Georgiev [16] | 2008 | 25 | 58 | 26 | 158 | 110 | 5 | 20 | 7 | 30 | 34 | 109/32 | Yes |
| Isbir [27] | 2003 | 37 | 49 | 14 | 123 | 77 | 20 | 40 | 12 | 80 | 64 | 100/72 | Yes |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, L.; Li, Y.-M. Lipoprotein Lipase (LPL) Polymorphism and the Risk of Coronary Artery Disease: A Meta-Analysis. Int. J. Environ. Res. Public Health 2017, 14, 84. https://doi.org/10.3390/ijerph14010084
Xie L, Li Y-M. Lipoprotein Lipase (LPL) Polymorphism and the Risk of Coronary Artery Disease: A Meta-Analysis. International Journal of Environmental Research and Public Health. 2017; 14(1):84. https://doi.org/10.3390/ijerph14010084
Chicago/Turabian StyleXie, Li, and You-Mei Li. 2017. "Lipoprotein Lipase (LPL) Polymorphism and the Risk of Coronary Artery Disease: A Meta-Analysis" International Journal of Environmental Research and Public Health 14, no. 1: 84. https://doi.org/10.3390/ijerph14010084





