Occurrence of Virulence Genes Associated with Human Pathogenic Vibrios Isolated from Two Commercial Dusky Kob (Argyrosmus japonicus) Farms and Kareiga Estuary in the Eastern Cape Province, South Africa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Bacteriological Analyses
2.3. Molecular Confirmation of Vibrio Species
2.4. Detection of Virulence Genes
2.5. Biotyping of V. Vulnificus
3. Results
3.1. Prevalence of Vibrio Species
3.2. Prevalence of Vibrio Virulence Genes
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Iwamoto, M.; Ayers, T.; Mahon, B.E.; Swerdlow, D.L. Epidemiology of seafood-associated infections in the United States. Clin. Microbiol. Rev. 2010, 23, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Eyisi, O.A.; Nwodo, U.U.; Iroegbu, C.U. Distribution of Vibrio species in shellfish and water samples collected from the atlantic coastline of south-east Nigeria. J. Health Popul. Nutr. 2013, 31, 314. [Google Scholar] [PubMed]
- Machado, A.; Bordalo, A.A. Detection and Quantification of Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus in Coastal Waters of Guinea-Bissau (West Africa). EcoHealth 2016, 13, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Chowdhury, G.; Pazhani, G.P.; Guin, S.; Dutta, S.; Ghosh, S.; Rajendran, K.; Nandy, R.K.; Mukhopadhyay, A.K.; Bhattacharya, M.K.; et al. Vibrio cholerae non-O1, non-O139 serogroups and cholera-like diarrhea, Kolkata, India. Emerg. Infect. Dis. 2013, 19, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.; Kendall, M.; Vugia, D.J.; Henao, O.L.; Mahon, B.E. Increasing rates of vibriosis in the United States, 1996–2010: Review of surveillance data from 2 systems. Clin. Infect. Dis. 2012, 54 (Suppl. S5), S391–S395. [Google Scholar] [CrossRef] [PubMed]
- Daniels, N.A.; MacKinnon, L.; Bishop, R.; Altekruse, S.; Ray, B.; Hammond, R.M.; Thompson, S.; Wilson, S.; Bean, N.H.; Griffin, P.M.; et al. Vibrio parahaemolyticus infections in the United States, 1973–1998. J. Infect. Dis. 2000, 181, 1661–1666. [Google Scholar] [CrossRef] [PubMed]
- Su, H.P.; Chiu, S.I.; Tsai, J.L.; Lee, C.L.; Pan, T.M. Bacterial food-borne illness outbreaks in northern Taiwan, 1995–2001. J. Infect. Chemother. 2005, 11, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Terzi Gulel, G.; Martinez-Urtaza, J. Molecular characterizations of Vibrio parahaemolyticus in seafood from the Black Sea, Turkey. Lett. Appl. Microbiol. 2016, 62, 494–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, M.; Deng, Y.; Zhang, X.; Liu, G.; Huang, Y.; Lin, C.; Li, J.; Yan, H.; Li, X.; Jia, L.; et al. Etiology of acute diarrhea due to enteropathogenic bacteria in Beijing, China. J. Infect. 2012, 65, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Hiyoshi, H.; Kodama, T.; Iida, T.; Honda, T. Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect. Immun. 2010, 78, 1772–1780. [Google Scholar] [CrossRef] [PubMed]
- Kushawaha, A.; Mobarakai, N.; Cooper, M.; Rose, K.; Awasum, M. Necrotising fasciitis with Vibrio vulnificus: A limb threatening dermatologic complication following exposure to marine life. BMJ Case Rep. 2010. [Google Scholar] [CrossRef] [PubMed]
- Bisharat, N.; Agmon, V.; Finkelstein, R.; Raz, R.; Ben-Dror, G.; Lerner, L.; Soboh, S.; Colodner, R.; Cameron, D.N.; Wykstra, D.L.; et al. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 1999, 354, 1421–1424. [Google Scholar] [CrossRef]
- Oliver, J.D. Wound infections caused by Vibrio vulnificus and other marine bacteria. Epidemiol. Infect. 2005, 133, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Vickery, M.C.; Nilsson, W.B.; Strom, M.S.; Nordstrom, J.L.; DePaola, A. A real-time PCR assay for the rapid determination of 16S rRNA genotype in Vibrio vulnificus. J. Microbiol. Methods 2007, 68, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Ramamurthy, T.; Chowdhury, G.; Pazhani, G.P.; Shinoda, S. Vibrio fluvialis: An emerging human pathogen. Vibrio Ecol. Pathog. Evolut. 2014, 5, 91. [Google Scholar] [CrossRef] [PubMed]
- Lagana, P.; Caruso, G.; Minutoli, E.; Zaccone, R.; Delia, S. Susceptibility to antibiotics of Vibrio spp. and Photobacterium damsela ssp. piscicida strains isolated from Italian aquaculture farms. New Microbiol. 2011, 34, 53–63. [Google Scholar] [PubMed]
- Igbinosa, E.O.; Okoh, A.I. Vibrio fluvialis: An unusual enteric pathogen of increasing public health concern. Int. J. Environ. Res. Public Health 2010, 7, 3628–3643. [Google Scholar] [CrossRef] [PubMed]
- Adebayo-Tayo, B.C.; Okonko, I.O.; Esen, C.U.; Odu, N.N.; Onoh, C.C.; Igwiloh, N.J.P. Incidence of potentially pathogenic Vibrio spp. in fresh seafood from Itu Creek in Uyo, Akwa Ibom State, Nigeria. World Appl. Sci. J. 2011, 15, 985–991. [Google Scholar]
- Waldor, M.K.; Mekalanos, J.J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 1996, 272, 1910. [Google Scholar] [CrossRef] [PubMed]
- Bina, X.R.; Taylor, D.L.; Vikram, A.; Ante, V.M.; Bina, J.E. Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo (Phe-Pro). mBio 2013, 4, e00366-13. [Google Scholar] [CrossRef] [PubMed]
- Terzi, G.; Büyüktanır, Ö.; Yurdusev, N. Detection of the tdh and trh genes in Vibrio parahaemolyticus isolates in fish and mussels from Middle Black Sea Coast of Turkey. Lett. Appl. Microbiol. 2009, 49, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.K.; Oliver, J.D. Vibrio vulnificus: Disease and pathogenesis. Infect. Immun. 2009, 77, 1723–1733. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, R.; Sinha, S.; Mukhopadhyay, A.K.; Asakura, M.; Yamasaki, S.; Bhattacharya, S.K.; Nair, G.B.; Ramamurthy, T. Species-specific identification of Vibrio fluvialis by PCR targeted to the conserved transcriptional activation and variable membrane tether regions of the toxR gene. J. Med. Microbiol. 2006, 55, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Cui, X.; Du, X.; Kan, B.; Liang, W. The virulence phenotypes and molecular epidemiological characteristics of Vibrio fluvialis in China. Gut Pathog. 2013, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Igbinosa, E.O.; Obi, L.C.; Okoh, A.I. Occurrence of potentially pathogenic vibrios in final effluents of a wastewater treatment facility in a rural community of the Eastern Cape Province of South Africa. Res. Microbiol. 2009, 160, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Igbinosa, E.O.; Obi, C.L.; Okoh, A.I. Seasonal abundance and distribution of Vibrio species in the treated effluent of wastewater treatment facilities in suburban and urban communities of Eastern Cape Province, South Africa. J. Microbiol. 2011, 49, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Okoh, A.I.; Sibanda, T.; Nongogo, V.; Adefisoye, M.; Olayemi, O.O.; Nontongana, N. Prevalence and characterisation of non-cholerae Vibrio spp. in final effluents of wastewater treatment facilities in two districts of the Eastern Cape Province of South Africa: Implications for public health. Environ. Sci. Pollut. Res. 2015, 22, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- American Public Health Association (APHA). Standard Methods for Examination of Water and Waste Water, 20th ed.; APHA-AWWA-WEF: Washington, DC, USA, 1998; pp. 9–56. [Google Scholar]
- Kwok, A.Y.; Wilson, J.T.; Coulthart, M.; Ng, L.K.; Mutharia, L.; Chow, A.W. Phylogenetic study and identification of human pathogenic Vibrio species based on partial hsp60 gene sequences. Can. J. Microbiol. 2002, 48, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Neogi, S.B.; Chowdhury, N.; Asakura, M.; Hinenoya, A.; Haldar, S.; Saidi, S.M.; Kogure, K.; Lara, R.J.; Yamasaki, S. A highly sensitive and specific multiplex PCR assay for simultaneous detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus. Lett. Appl. Microbiol. 2010, 51, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.K.; Ponmariappan, S.; Kamboj, D.V.; Singh, L. Single multiplex polymerase chain reaction for environmental surveillance of toxigenic—Pathogenic O1 and non-O1Vibrio cholerae. Folia Microbiol. 2007, 52, 81–85. [Google Scholar] [CrossRef]
- Kim, Y.B.; Okuda, J.; Matsumoto, C.; Takahashi, N.; Hashimoto, S.; Nishibuchi, M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 1999, 37, 1173–1177. [Google Scholar] [PubMed]
- Rosche, T.M.; Yano, Y.; Oliver, J.D. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immun. 2005, 49, 381–389. [Google Scholar] [CrossRef]
- Zaidenstein, R.; Sadik, C.; Lerner, L.; Valinsky, L.; Kopelowitz, J.; Yishai, R.; Agmon, V.; Parsons, M.; Bopp, C.; Weinberger, M. Clinical characteristics and molecular subtyping of Vibrio vulnificus illnesses, Israel. Emerg. Infect. Dis. 2008, 14, 1875–1882. [Google Scholar] [CrossRef] [PubMed]
- Huss, H.H. Control of indigenous pathogenic bacteria in seafood. Food Control 1997, 8, 91–98. [Google Scholar] [CrossRef]
- Igbinosa, E.O. Detection and antimicrobial resistance of Vibrio isolates in aquaculture environments: Implications for public health. Microb. Drug Resist. 2016, 22, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Nsofor, C.A.; Kemajou, S.T.; Nsofor, C. MIncidence and antibiotic susceptibility pattern of Vibrio species isolated from sea foods sold in Port-Harcourt, Nigeria. Afr. J. Bacteriol. Res. 2014, 6, 13–16. [Google Scholar]
- Furniss, A.L.; Lee, J.V.; Donovan, T.J. Group F, a new Vibrio? Lancet 1977, 310, 565–566. [Google Scholar] [CrossRef]
- Kobayashi, K.; Ohnaka, T. Food poisoning due to newly recognized pathogens. Asian Med. J. 1989, 32, 1–12. [Google Scholar]
- Thekdi, R.J.; Lakhani, A.G.; Rale, V.B.; Panse, M.V. An outbreak of food poisoning suspected to be caused by Vibrio fluvialis. J. Diarrhoeal Dis. Res. 1990, 8, 163–165. [Google Scholar] [PubMed]
- Levine, W.C.; Griffin, P.M. Gulf Coast Vibrio Working Group. Vibrio infections on the Gulf Coast: Results of first year of regional surveillance. J. Infect. Dis. 1993, 167, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.H.; Hwang, C.K.; Chin, C.; Lin, H.H.; Wong, W.W.; Liu, C.Y. Severe watery diarrhoea and bacteraemia caused by Vibrio fluvialis. J. Infect. 2006, 52, e95–e98. [Google Scholar] [CrossRef] [PubMed]
- Miron, D.; Lev, A.; Colodner, R.; Merzel, Y. Vibrio vulnificus necrotizing fasciitis of the calf presenting with compartment syndrome. Pediatr. Infect. Dis. J. 2003, 22, 666–668. [Google Scholar] [CrossRef] [PubMed]
- Mead, P.S.; Slutsker, L.; Dietz, V.; McCaig, L.F.; Bresee, J.S.; Shapiro, C.; Griffin, P.M.; Tauxe, R.V. Food-related illness and death in the United States. Emerg. Infect. Dis. 1999, 5, 607. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.G.; Acheson, D. Cholera and other types of vibriosis: A story of human pandemics and oysters on the half shell. Clin. Infect. Dis. 2003, 37, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.C.; Lui, S.H.; Wang, T.K.; Lee, C.L.; Chiou, C.S.; Lui, D.P.; Nishibuchi, M.; Lee, B.K. Characterisation of Vibrio paraheamolyticus O3:K6 from Asia. App. Environ. Microbiol. 2000, 66, 3981–3986. [Google Scholar] [CrossRef]
- Zulkifli, Y.; Alitheen, N.B.; Son, R.; Yeap, S.K.; Lesly, M.B.; Raha, A.R. Identification of Vibrio parahaemolyticus isolates by PCR targeted to the ToxR gene and detection of virulence genes. Int. Food Res. J. 2009, 16, 289–296. [Google Scholar]
- Koralage, M.S.G.; Alter, T.; Pichpol, D.; Strauch, E.; Zessin, K.H.; Huehn, S. Prevalence and molecular characteristics of Vibrio spp. isolated from preharvest shrimp of the North Western Province of Sri Lanka. J. Food Prot. 2012, 75, 1846–1850. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.L.; Lüdeke, C.H.; Bowers, J.C.; Garrett, N.; Fischer, M.; Parsons, M.B.; Bopp, C.A.; DePaola, A. Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xie, X.; Shi, X.; Lin, Y.; Qiu, Y.; Mou, J.; Chen, Q.; Lu, Y.; Zhou, L.; Jiang, M.; et al. Vibrio parahaemolyticus, Southern Coastal Region of China, 2007–2012. Emerg. Infect. Dis. 2014, 20, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Pazhani, G.P.; Bhowmik, S.K.; Ghosh, S.; Guin, S.; Dutta, S.; Rajendran, K.; Saha, D.R.; Nandy, R.K.; Bhattacharya, M.K.; Mukhopadhyay, A.K.; et al. Trends in the epidemiology of pandemic and non-pandemic strains of Vibrio parahaemolyticus isolated from diarrheal patients in Kolkata, India. PLoS Negl. Trop. Dis. 2014, 8, e2815. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, J.C.; Gerding, M.J.; Jones, S.H.; Whistler, C.A. Comparison of the pathogenic potentials of environmental and clinical Vibrio parahaemolyticus strains indicates a role for temperature regulation in virulence. Appl. Environ. Microbiol. 2010, 76, 7459–7465. [Google Scholar] [CrossRef] [PubMed]
- Raghunath, P. Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 2015, 5, 805. [Google Scholar] [CrossRef] [PubMed]
- Gulig, P.A.; Bourdage, K.L.; Starks, A.M. Molecular pathogenesis of Vibrio vulnificus. J. Microbiol. 2005, 43 (Suppl. S1), 118–131. [Google Scholar] [PubMed]
- Bier, N.; Diescher, S.; Strauch, E. Multiplex PCR for detection of virulence markers of Vibrio vulnificus. Lett. Appl. Microbiol. 2015, 60, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Warner, E.; Oliver, J.D. Population structures of two genotypes of Vibrio vulnificus in oysters (Crassostrea virginica) and seawater. Appl. Environ. Microbiol. 2008, 74, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, A.; Gómez Gil Rodríguez, B.; Wong-Chang, I.; Lizárraga-Partida, M.L. Genetic characterization of Vibrio vulnificus strains isolated from oyster samples in Mexico. Int. J. Environ. Health Res. 2015, 25, 614–627. [Google Scholar] [CrossRef] [PubMed]
- Amaro, C.; Biosca, E.G. Vibrio vulnificus biotype 2, pathogenic for eels, is also an opportunistic pathogen for humans. Appl. Environ. Microbiol. 1996, 62, 1454–1457. [Google Scholar] [PubMed]
- Sanjuán, E.; Amaro, C. Multiplex PCR assay for detection of Vibrio vulnificus biotype 2 and simultaneous discrimination of serovar E strains. Appl. Environ. Microbiol. 2007, 73, 2029–2032. [Google Scholar] [CrossRef] [PubMed]

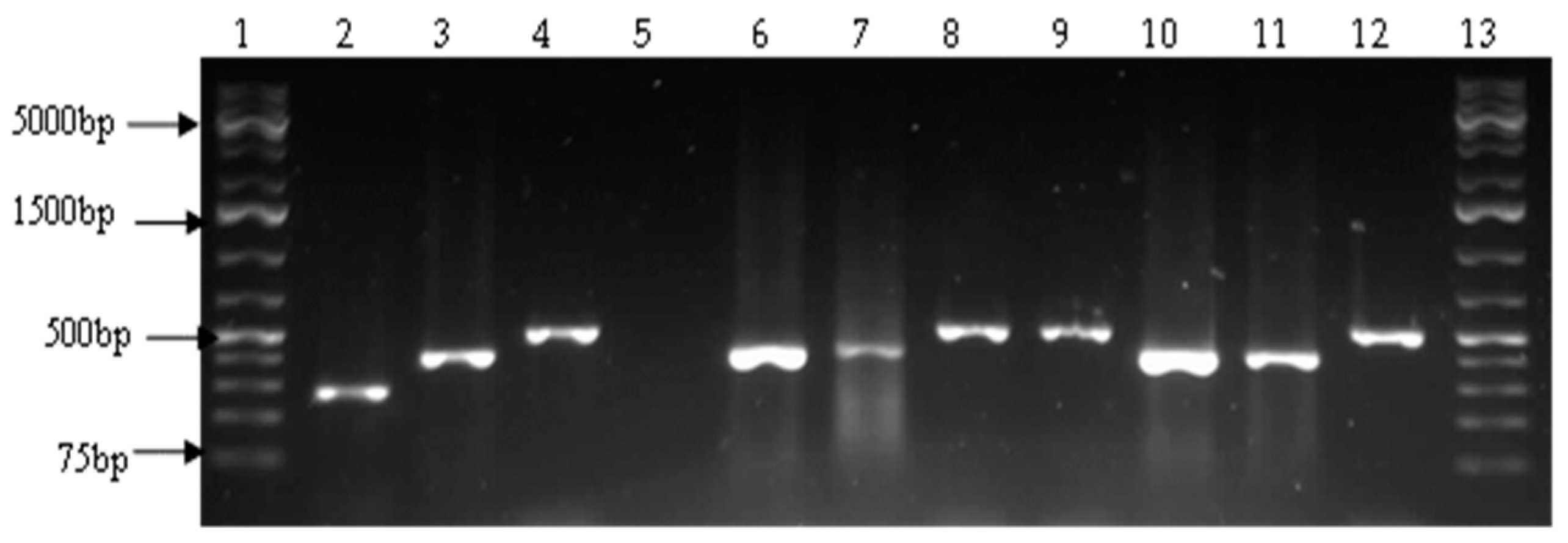
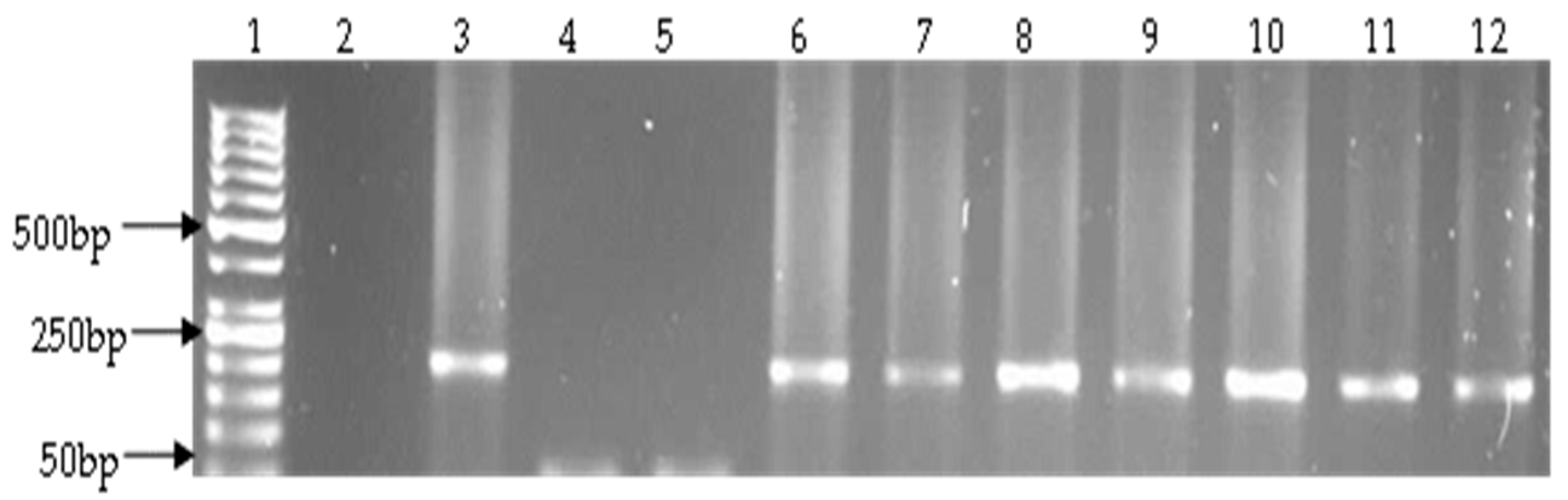

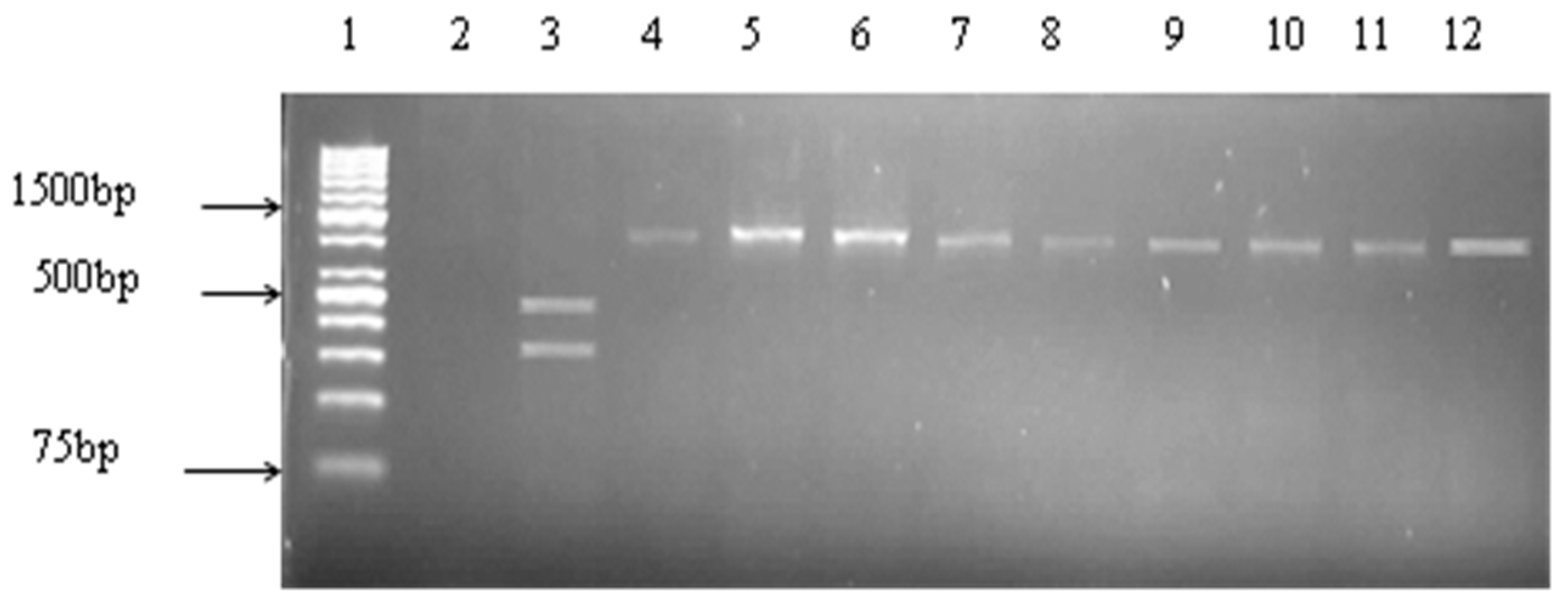
| Target Species | Target Gene | Primer Sequence (5′–3′) | Amplicon Size (bp) | Cycling Conditions | References |
|---|---|---|---|---|---|
| Vibrio | 16S rRNA | CGGTGAAATGCGTAGAGAT TTACTAGCGATTCCGAGTTC | 663 | Initial denaturation at 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 52 °C for 30 s and 72 °C for 60 s. Final extension was at 72 °C for 10 min. | [29] |
| V. parahaemolyticus | toxR | TGTACTGTTGAACGCCTAA CACGTTCTCATACGAGTG | 503 | Initial denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s. A final extension step at 72 °C for 10 min. | [30] |
| V. vulnificus | vvhA | ACTCAACTATCGTGCACG ACACTGTTCGACTGTGAG | 366 | ||
| V. cholerae | toxR | GAAGCTGCTCATGACATC AAGATCAGGGTGGTTATTC | 275 | ||
| OmpW | CACCAAGAAGGTGACTTTATTGTG GGTTTGTCGAATTAGCTTCACC | 304 | Initial denaturation at 94 °C for 10 min, followed by 30 cycles at 94 °C for 60 s, 59 °C for 60 s and 72 °C for 2 min. Final elongation step at 72 °C for 10 min. | [31] | |
| V. fluvialis | toxR | GGATACGGCACTTGAGTAAGACTC GACCAGGGCTTTGAGGTGGACGAC | 217 | Initial denaturation step at 94 °C for 5 min, followed by 30 cycles at 94 °C for 30 s, 57 °C for 60 s and 72 °C for 90 s. A final extension step at 72 °C for 7 min. | [23] |
| Primer | Sequence (5′–3′) | Product Size (bp) | Cycling Conditions | References |
|---|---|---|---|---|
| tdhF tdhR | GGTCTAAATGGCTGACATC CCACTACCACTCTCATATGC | 199 | Initial denaturation at 96 °C for 5 min, followed by 35 cycles of 94 °C for 60 s, 55 °C for 60 s and 72 °C for 60 s. Final extension at 72 °C for 7 min. | [32] |
| trhF trhR | CATTTCCGCTCTCATATGC GGCTCAAAATGGTTAAGCG | 250 | ||
| vcgCP1 vcgP3 | AGCTGCCGATAGCGATCT CGCTTAGGATGATCGGTG | 278 | Initial denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 40 s, 56°C for 40 s and 72 °C for 60 s. Final extension at 72 °C for 7 min. | [33] |
| vcgEP2 vcgP3 | CTCAATTGACAATGATCT CGCTTAGGATGATCGGTG | 278 | Initial denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 40 s, 49 °C for 40 s and 72 °C for 60 s. Final extension at 72 °C for 7 min. | [33] |
| vfh-F vfh-R | GCGCGTCAGTGGTGGTGAAG TCGGTCGAACCGCTCTCGCTT | 800 | Initial denaturation at 93 °C for 15 min, followed by 35 cycles at 92 °C for 40 s, 50–62 °C for 60 s and 72 °C for 90 s. Final elongation step at 72 °C for 7 min. | [24] |
| hupO-F hupO-R | ATTACGCACAACGAGTCGAAC ATTGAGATGGT AAACAGCGCC | 600 | ||
| vfpA-F vfpA-R | TACAACGTCAAGTTAAAGGC GTAGGCGCTGTAGCCTTTCA | 1790 | ||
| stn-F stn-R | GGTGCAACATAATAAACAGTCAACAA TAGTGGTATGCGTTGCCAGC | 375 |
| Sampling Site | Source (n = No. of Vibrio Positives by PCR) | Vibrio Species | Total | ||
|---|---|---|---|---|---|
| V. fluvialis | V. parahaemolyticus | V. vulnificus | |||
| Kariega Estuary | Water (n = 39) | 3 (7.69%) | 4 (10.26%) | 0 (0.00%) | 7 (17.95%) |
| Fish (n = 57) | 7 (12.28%) | 5 (8.77%) | 1 (1.75%) | 13 (22.81%) | |
| Farm 1 | Water (n = 73) | 54 (73.97%) | 4 (5.48%) | 2 (2.74%) | 60 (82.19%) |
| Fish (n = 290) | 109 (37.59%) | 14 (11.93%) | 61 (55.96%) | 184 (63.45%) | |
| Farm 2 | Water (n = 45) | 5 (11.11%) | 3 (6.67%) | 5 (11.11%) | 13 (28.89%) |
| Fish (n = 102) | 15 (14.71%) | 3 (2.94%) | 5 (4.91%) | 23 (22.55%) | |
| Total | 606 | 193 (31.85%) | 33 (5.45%) | 74 (12.21%) | 300 (49.50%) |
| Site | V. fluvialis Virulence Genes | |||
|---|---|---|---|---|
| vfh | hupO | stn | vfpA | |
| Farm 1 (n = 10) | 2 (20%) | 2 (20%) | 2 (20%) | 0 (0%) |
| Farm 2 (n = 163) | 10 (6.1%) | 18 (11%) | 21 (12.9%) | 0 (0%) |
| Kareiga Estuary (n = 20) | 0 (%) | 0 (0%) | 3 (15%) | 0 (0%) |
| Total (n = 193) | 12 (6.2%) | 20 (10.4%) | 26 (13.5%) | 0 (0%) |
| Isolate Code | V. fluvialis Virulence Genes | ||
|---|---|---|---|
| vfh | hupO | stn | |
| 11 | + | ||
| 30 | + | + | |
| 36 | + | + | + |
| 57 | + | ||
| 58 | + | + | |
| 72 | + | ||
| 206a | + | + | |
| 207d | + | ||
| 225b | + | + | |
| 231a | + | + | |
| 241a | + | + | |
| 241d | + | + | |
| 241g | + | + | |
| 241j | + | + | |
| 252g | + | ||
| 253a | + | + | |
| 253b | + | + | |
| 253c | + | + | |
| 253d | + | + | |
| 262d | + | ||
| 271a | + | + | + |
| 271b | + | + | + |
| 276b | + | + | |
| 276f | + | + | |
| 277d | + | ||
| 278a | + | + | |
| 278c | + | ||
| 289 | + | + | |
| 308b | + | ||
| 308f | + | ||
| 309a | + | ||
| 330h | + | ||
| 369b | + | ||
| 375a | + | ||
| 379a | + | ||
| Total (n = 35) | 12 | 20 | 26 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fri, J.; Ndip, R.N.; Njom, H.A.; Clarke, A.M. Occurrence of Virulence Genes Associated with Human Pathogenic Vibrios Isolated from Two Commercial Dusky Kob (Argyrosmus japonicus) Farms and Kareiga Estuary in the Eastern Cape Province, South Africa. Int. J. Environ. Res. Public Health 2017, 14, 1111. https://doi.org/10.3390/ijerph14101111
Fri J, Ndip RN, Njom HA, Clarke AM. Occurrence of Virulence Genes Associated with Human Pathogenic Vibrios Isolated from Two Commercial Dusky Kob (Argyrosmus japonicus) Farms and Kareiga Estuary in the Eastern Cape Province, South Africa. International Journal of Environmental Research and Public Health. 2017; 14(10):1111. https://doi.org/10.3390/ijerph14101111
Chicago/Turabian StyleFri, Justine, Roland Ndip Ndip, Henry Akum Njom, and Anna Maria Clarke. 2017. "Occurrence of Virulence Genes Associated with Human Pathogenic Vibrios Isolated from Two Commercial Dusky Kob (Argyrosmus japonicus) Farms and Kareiga Estuary in the Eastern Cape Province, South Africa" International Journal of Environmental Research and Public Health 14, no. 10: 1111. https://doi.org/10.3390/ijerph14101111




