Genotypic and Epidemiological Trends of Acute Gastroenteritis Associated with Noroviruses in China from 2006 to 2016
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Case Definition and Outbreak Definition
- (1)
- A laboratory-confirmed case of NoV infection is defined as a patient with three episodes of loose stools and/or two episodes of vomiting within 24 h, with detection of NoV in stool, rectal swabs or vomitus specimens by Reverse Transcription-Polymerase Chain Reaction (RT-PCR) nucleic acid or enzyme-linked immunosorbent assay (ELISA) antigen testing. RT-PCR and ELISA protocols showed in http://www.chinacdc.cn/jkzt/crb/qt/nrbdjxwcy/jszl_2273/201511/t2015 1120_122120.html.
- (2)
- A clinical diagnosed NoV case is defined as a patient with three episodes of loose stools and/or two episodes of vomiting within 24 h, and with an epidemiologic link to laboratory-confirmed cases in one outbreak.
- (3)
- An outbreak of NoV infection should satisfy the following criteria: first, the event reports 20 or more NoVs cases in 7 days; second, the event occurs in one setting, for example: a school, kindergarten, hospital, nursing home, etc.; third, the reported cases have the same exposure from a common source of infection or by person-to-person transmission; and fourth, two or more cases are laboratory-confirmed cases.
2.3. Data Source
2.3.1. Outbreak Information
2.3.2. Sequences Information
2.4. Data Analyses
3. Results
3.1. Epidemiological Characteristics of NoVs Infection Outbreaks in China
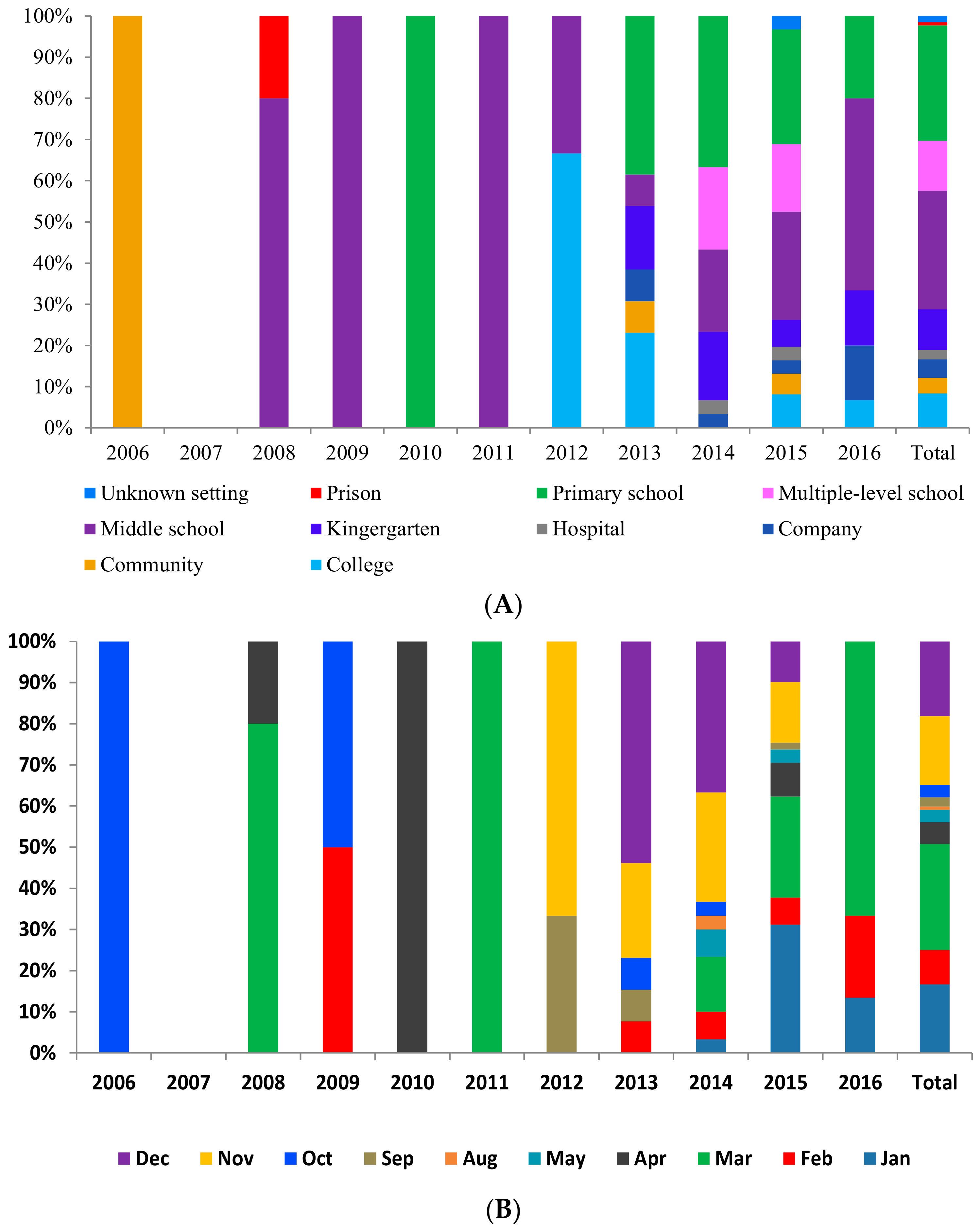
3.1.1. Geographic Distribution
3.1.2. Temporal Distribution
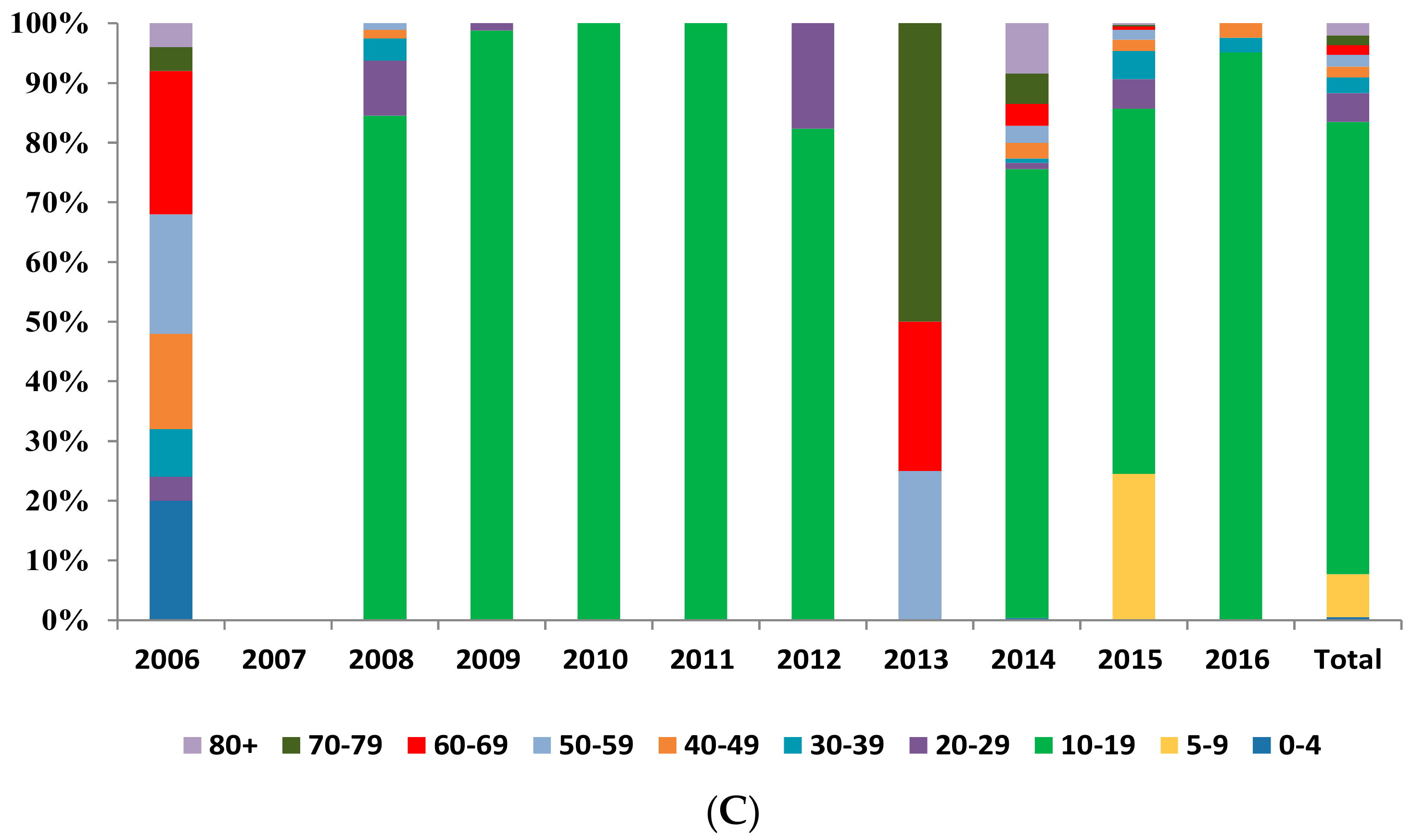
3.1.3. Population Distribution
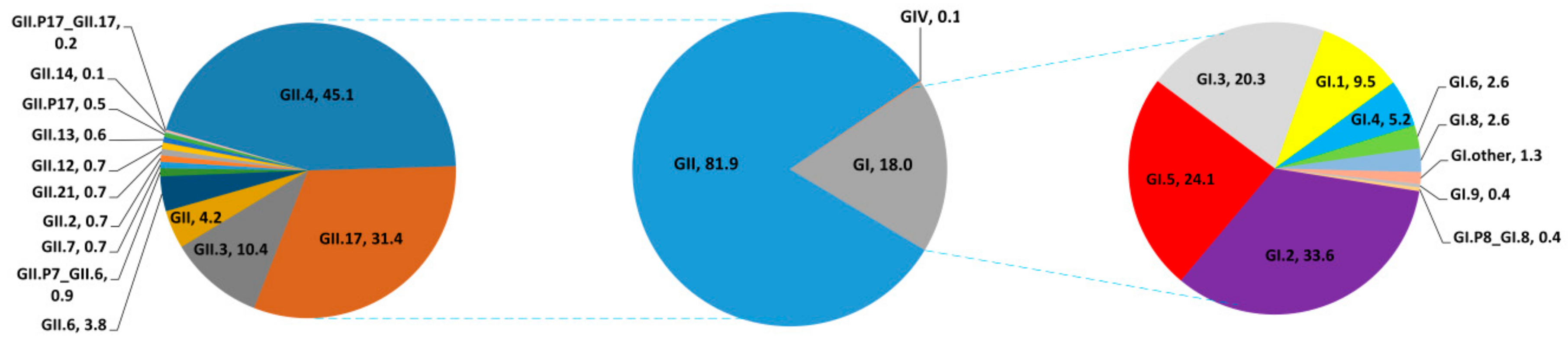
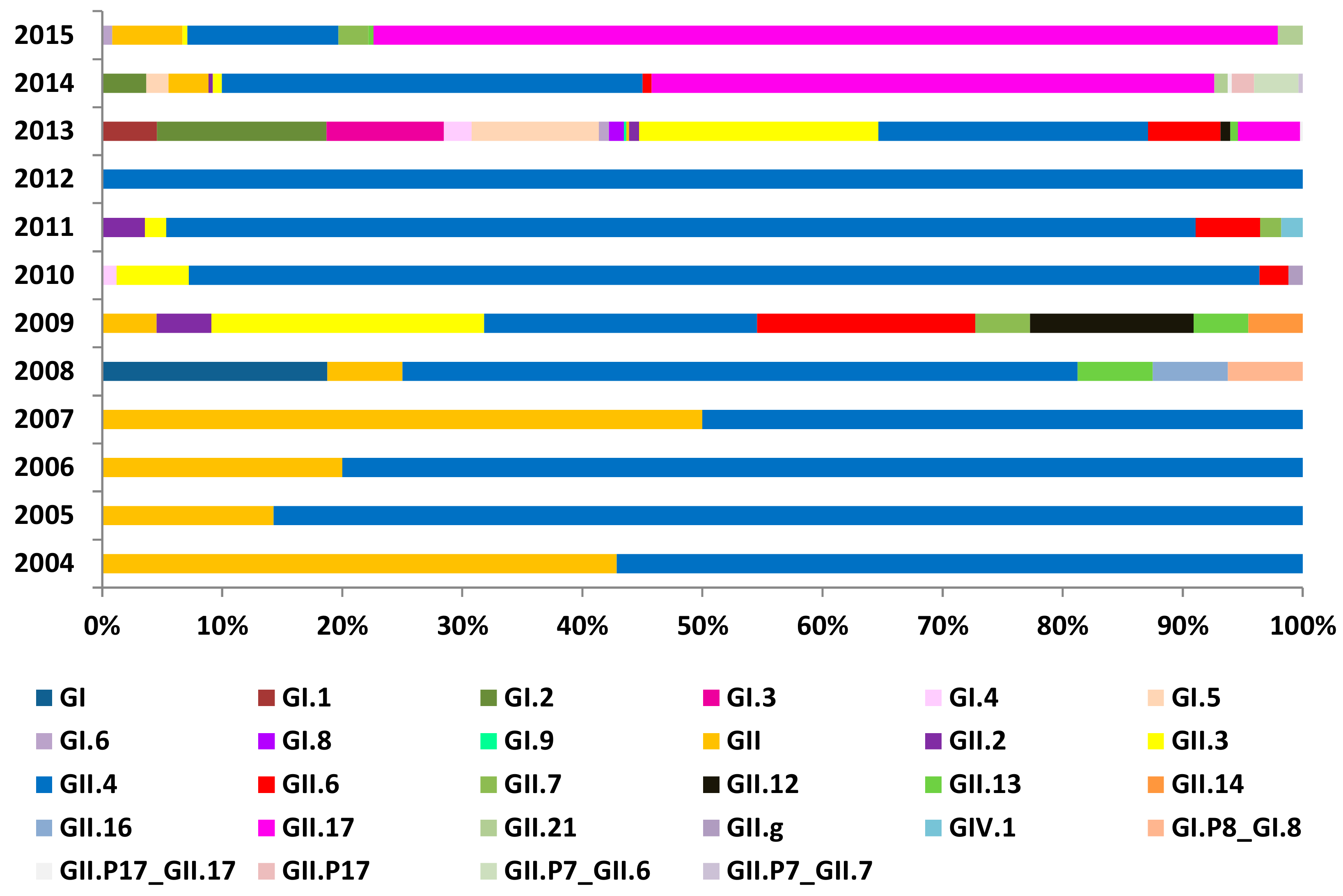
3.2. Genotype Distribution of NoVs Infection in China
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| NoVs | Noroviruses |
| RNA | Ribonucleic acid |
| ORF | Open reading frames |
| NS | Nonstructural mature proteins |
| NNDRS | National Notifiable Disease Reporting System |
| CDC | Center for Disease Control and Prevention |
References
- Matthews, J.E.; Dickey, B.W.; Miller, R.D.; Felzer, J.R.; Dawson, B.P.; Lee, A.S.; Rocks, J.J.; Kiel, J.; Montes, J.S.; Moe, C.L.; et al. The epidemiology of published norovirus outbreaks: A review of risk factors associated with attack rate and genogroup. Epidemiol. Infect. 2012, 140, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Vinje, J. Advances in laboratory methods for detection and typing of norovirus. J. Clin. Microbiol. 2015, 53, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.P.; Ando, T.; Fankhauser, R.L.; Beard, R.S.; Glass, R.I.; Monroe, S.S. Norovirus classification and proposed strain nomenclature. Virology 2006, 346, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Takano, T.; Kusuhara, H.; Kuroishi, A.; Takashina, M.; Doki, T.; Nishinaka, T.; Hohdatsu, T. Molecular characterization and pathogenicity of a genogroup GVI feline norovirus. Vet. Microbiol. 2015, 178, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.J.; Trainor, E.; Callaghan, A.; O’Brien, S.J.; Cunliffe, N.A.; Iturriza-Gomara, M. Early detection of epidemic GII-4 norovirus strains in UK and Malawi: Role of surveillance of sporadic acute gastroenteritis in anticipating global epidemics. PLoS ONE 2016, 11, e0146972. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Jeong, H.J.; Hwang, B.M.; Yoo, C.K.; Chung, G.T.; Jeong, H.; Kang, Y.H.; Lee, D.Y. Epidemics of norovirus GII.4 variant in outbreak cases in Korea, 2004–2012. Osong Public Health Res. Perspect. 2015, 6, 318–321. [Google Scholar] [CrossRef] [PubMed]
- Vega, E.; Barclay, L.; Gregoricus, N.; Shirley, S.H.; Lee, D.; Vinje, J. Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013. J. Clin. Microbiol. 2014, 52, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Division of Viral Diseases; National Center for Immunization and Respiratory Diseases; Centers for Disease Control and Prevention. Updated norovirus outbreak management and disease prevention guidelines. MMWR: Recomm. Rep. 2011, 60, 1–18. [Google Scholar]
- Robilotti, E.; Deresinski, S.; Pinsky, B.A. Norovirus. Clin. Microbiol. Rev. 2015, 28, 134–164. [Google Scholar] [CrossRef] [PubMed]
- Kapikian, A.Z.; Wyatt, R.G.; Dolin, R.; Thornhill, T.S.; Kalica, A.R.; Chanock, R.M. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 1972, 10, 1075–1081. [Google Scholar] [PubMed]
- Glass, R.I.; Parashar, U.D.; Estes, M.K. Norovirus gastroenteritis. N. Engl. J. Med. 2009, 361, 1776–1785. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.C.; Vinje, J.; Szilagyi, P.G.; Edwards, K.M.; Staat, M.A.; Weinberg, G.A.; Hall, C.B.; Chappell, J.; Bernstein, D.I.; Curns, A.T.; et al. Norovirus and medically attended gastroenteritis in U.S. Children. N. Engl. J. Med. 2013, 368, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.M.; Hall, A.J.; Robinson, A.E.; Verhoef, L.; Premkumar, P.; Parashar, U.D.; Koopmans, M.; Lopman, B.A. Global prevalence of norovirus in cases of gastroenteritis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 725–730. [Google Scholar] [CrossRef]
- Lopman, B. Global Burden of Norovirus and Prospects for Vaccine Development; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2017. [Google Scholar]
- Patel, M.M.; Hall, A.J.; Vinje, J.; Parashar, U.D. Noroviruses: A comprehensive review. J. Clin. Virol. 2009, 44, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Manso, C.F.; Romalde, J.L. Molecular epidemiology of norovirus from patients with acute gastroenteritis in northwestern Spain. Epidemiol. Infect. 2015, 143, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Newman, K.L.; Moe, C.L.; Kirby, A.E.; Flanders, W.D.; Parkos, C.A.; Leon, J.S. Norovirus in symptomatic and asymptomatic individuals: Cytokines and viral shedding. Clin. Exp. Immunol. 2016, 184, 347–357. [Google Scholar] [CrossRef] [PubMed]
- White, P.A. Evolution of norovirus. Clin. Microbiol. Infect. 2014, 20, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Pringle, K.; Lopman, B.; Vega, E.; Vinje, J.; Parashar, U.D.; Hall, A.J. Noroviruses: Epidemiology, immunity and prospects for prevention. Future Microbiol. 2015, 10, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Pan, H.; Hu, J.; Wu, H.; Li, J.; Xiao, W.; Zhang, X.; Yuan, Z.; Wu, F. Epidemiology of norovirus infections among diarrhea outpatients in a diarrhea surveillance system in Shanghai, China: A cross-sectional study. BMC Infect. Dis. 2015, 15, 183. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Zhang, H.; Lin, X.; Hou, P.; Wang, S.; Tao, Z.; Bi, Z.; Xu, A. A waterborne norovirus gastroenteritis outbreak in a school, Eastern China. Epidemiol. Infect. 2016, 144, 1212–1219. [Google Scholar] [CrossRef] [PubMed]
- Lindesmith, L.C.; Beltramello, M.; Donaldson, E.F.; Corti, D.; Swanstrom, J.; Debbink, K.; Lanzavecchia, A.; Baric, R.S. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog. 2012, 8, e1002705. [Google Scholar] [CrossRef] [PubMed]
- Kroneman, A.; Verhoef, L.; Harris, J.; Vennema, H.; Duizer, E.; Van Duynhoven, Y.; Gray, J.; Iturriza, M.; Böttiger, B.; Falkenhorst, G.; et al. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the foodborne viruses in Europe network from 1 July 2001 to 30 June 2006. J. Clin. Microbiol. 2008, 46, 2959–2965. [Google Scholar] [CrossRef] [PubMed]
- Greer, A.L.; Drews, S.J.; Fisman, D.N. Why “winter” vomiting disease? Seasonality, hydrology, and norovirus epidemiology in Toronto, Canada. EcoHealth 2009, 6, 192–199. [Google Scholar] [CrossRef] [PubMed]
- De la Noue, A.C.; Estienney, M.; Aho, S.; Perrier-Cornet, J.M.; de Rougemont, A.; Pothier, P.; Gervais, P.; Belliot, G. Absolute humidity influences the seasonal persistence and infectivity of human norovirus. Appl. Environ. Microbiol. 2014, 80, 7196–7205. [Google Scholar] [CrossRef] [PubMed]
- Mounts, A.W.; Ando, T.; Koopmans, M.; Bresee, J.S.; Noel, J.; Glass, R.I. Cold weather seasonality of gastroenteritis associated with norwalk-like viruses. J. Infect. Dis. 2000, 181 (Suppl. 2), S284–S287. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Lin, J.; Gao, J.; Yao, W.; Wen, D.; Liu, G.; Han, J.; Ma, H.; Zhang, L.; Zhu, B. A norovirus-borne outbreak caused by contaminated bottled spring water in a school, Zhejiang province. Chin. J. Epidemiol. 2011, 32, 800–803. [Google Scholar]
- Xue, L.; Dong, R.; Wu, Q.; Li, Y.; Cai, W.; Kou, X.; Zhang, J.; Guo, W. Molecular epidemiology of noroviruses associated with sporadic gastroenteritis in Guangzhou, China, 2013–2015. Arch. Virol. 2016, 161, 1377–1384. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.; Deng, L.; Wang, M.; Li, X.; Ma, Y.; Liu, W. High prevalence and genetic diversity of noroviruses among children with sporadic acute gastroenteritis in Nanning city, China, 2010–2011. J. Med. Virol. 2015, 87, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Cai, W.; Wu, Q.; Kou, X.; Zhang, J.; Guo, W. Comparative genome analysis of a norovirus GII.4 strain GZ2013-L10 isolated from South China. Virus Genes 2016, 52, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Thongprachum, A.; Khamrin, P.; Maneekarn, N.; Hayakawa, S.; Ushijima, H. Epidemiology of gastroenteritis viruses in Japan: Prevalence, seasonality, and outbreak. J. Med. Virol. 2016, 88, 551–570. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.W.; Chu, C. Norovirus outbreaks occurred in different settings in the Republic of Korea. Osong Public Health Res. Perspect. 2015, 6, 281–282. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.J.; Wikswo, M.E.; Manikonda, K.; Roberts, V.A.; Yoder, J.S.; Gould, L.H. Acute gastroenteritis surveillance through the national outbreak reporting system, United States. Emerg. Infect. Dis. 2013, 19, 1305–1309. [Google Scholar] [CrossRef] [PubMed]
- Barret, A.S.; Jourdan-da Silva, N.; Ambert-Balay, K.; Delmas, G.; Bone, A.; Thiolet, J.M.; Vaillant, V. Surveillance for outbreaks of gastroenteritis in elderly long-term care facilities in France, November 2010 to May 2012. Euro Surveill. 2014, 19, 15–22. [Google Scholar] [CrossRef]
- Loury, P.; Le Guyader, F.S.; Le Saux, J.C.; Ambert-Balay, K.; Parrot, P.; Hubert, B. A norovirus oyster-related outbreak in a nursing home in France, January 2012. Epidemiol. Infect. 2015, 143, 2486–2493. [Google Scholar] [CrossRef] [PubMed]
- Friesema, I.H.; Vennema, H.; Heijne, J.C.; de Jager, C.M.; Morroy, G.; van den Kerkhof, J.H.; de Coster, E.J.; Wolters, B.A.; ter Waarbeek, H.L.; Fanoy, E.B.; et al. Norovirus outbreaks in nursing homes: The evaluation of infection control measures. Epidemiol. Infect. 2009, 137, 1722–1733. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Sun, J.; Chang, Z.; Li, H.; Liu, N.; Zhang, Q.; Zhang, J.; Wang, Z.; Duan, Z. Outbreaks of noroviral gastroenteritis and their molecular characteristics in China, 2006–2007. Chin. J. Epidemiol. 2010, 31, 549–553. [Google Scholar]
- Sang, S.; Zhao, Z.; Suo, J.; Xing, Y.; Jia, N.; Gao, Y.; Du, M.; Xie, L.; Deng, C.; Ren, S.; et al. Characteristics of outbreak and epidemiology of norovirus gastroenteritis in China. Chin. J. Nosocomiol. 2011, 21, 4245–4247. [Google Scholar]
- Lim, K.L.; Hewitt, J.; Sitabkhan, A.; Eden, J.S.; Lun, J.; Levy, A.; Merif, J.; Smith, D.; Rawlinson, W.D.; White, P.A. A multi-site study of norovirus molecular epidemiology in australia and New Zealand, 2013–2014. PLoS ONE 2016, 11, e0145254. [Google Scholar] [CrossRef] [PubMed]
- James, A.; Magnus, L.; Nancy, N.; Ferdinand, T.; Theresa, N.-A.; Tomas, B. Enteric viruses in healthy children in cameroon: Viral load and genotyping of norovirus strains. J. Med. Virol. 2011, 83, 2135–2142. [Google Scholar]
- Ha, S.; Choi, I.S.; Choi, C.; Myoung, J. Infection models of human norovirus: Challenges and recent progress. Arch. Virol. 2016, 161, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Zhirakovskaia, E.V.; Tikunov, A.Y.; Bodnev, S.A.; Klemesheva, V.V.; Netesov, S.V.; Tikunova, N.V. Molecular epidemiology of noroviruses associated with sporadic gastroenteritis in children in novosibirsk, Russia, 2003–2012. J. Med. Virol. 2015, 87, 740–753. [Google Scholar] [CrossRef] [PubMed]
- Rouhani, S.; Penataro Yori, P.; Paredes Olortegui, M.; Siguas Salas, M.; Rengifo Trigoso, D.; Mondal, D.; Bodhidatta, L.; Platts-Mills, J.; Samie, A.; Kabir, F.; et al. Norovirus infection and acquired immunity in 8 countries: Results from the MAL-ED study. Clin. Infect. Dis. 2016, 62, 1210–1217. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Jin, M.; Chen, K.; Zhang, H.; Yang, H.; Zhuo, F.; Zhao, D.; Zeng, H.; Yao, X.; Zhang, Z.; et al. Gastroenteritis outbreaks associated with the emergence of the new GII.4 sydney norovirus variant during the epidemic of 2012/13 in Shenzhen city, China. PLoS ONE 2016, 11, e0165880. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.G.; Ai, J.; Qi, X.; Zhang, J.; Tang, F.Y.; Zhu, Y.F. Emergence of two novel norovirus genotype II.4 variants associated with viral gastroenteritis in China. J. Med. Virol. 2014, 86, 1226–1234. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.G.; Park, P.H.; Lee, S.G.; Kim, J.E.; Kim, K.A.; Lee, H.K.; Park, E.M.; Park, M.K.; Jung, S.Y.; Lee, D.Y.; et al. Emergence of norovirus GII.4 variants in acute gastroenteritis outbreaks in South Korea between 2006 and 2013. J. Clin. Virol. 2015, 72, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.T.; Chen, H.C.; Yen, C.; Wu, C.Y.; Katayama, K.; Park, Y.; Hall, A.J.; Vinje, J.; Huang, J.C.; Wu, H.S. Epidemiology and molecular characteristics of norovirus GII.4 Sydney outbreaks in Taiwan, January 2012–December 2013. J. Med. Virol. 2015, 87, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shen, Z.; Zhu, Z.; Zhang, W.; Chen, H.; Qian, F.; Chen, H.; Wang, G.; Wang, M.; Hu, Y.; et al. Genotype distribution of norovirus around the emergence of Sydney_2012 and the antigenic drift of contemporary GII.4 epidemic strains. J. Clin. Virol. 2015, 72, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Debbink, K.; Lindesmith, L.C.; Donaldson, E.F.; Costantini, V.; Beltramello, M.; Corti, D.; Swanstrom, J.; Lanzavecchia, A.; Vinje, J.; Baric, R.S. Emergence of new pandemic GII.4 Sydney norovirus strain correlates with escape from herd immunity. J. Infect. Dis. 2013, 208, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Mai, H.; Gao, Y.; Pan, X.; Han, J.; Cong, X.; Wei, L. Study on norovirus Gii.4/Sydney 2012 variant in China. Chin. J. Epidemiol. 2014, 35, 157–162. [Google Scholar]
- Wang, H.B.; Wang, Q.; Zhao, J.H.; Tu, C.N.; Mo, Q.H.; Lin, J.C.; Yang, Z. Complete nucleotide sequence analysis of the norovirus GII.17: A newly emerging and dominant variant in China, 2015. Infect. Genet. Evol. 2016, 38, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Dinu, S.; Nagy, M.; Negru, D.G.; Popovici, E.D.; Zota, L.; Oprisan, G. Molecular identification of emergent GII.P17–GII.17 norovirus genotype, Romania, 2015. Euro Surveill. 2016, 21, 30141. [Google Scholar] [CrossRef] [PubMed]
- Parra, G.I.; Green, K.Y. Genome of emerging norovirus GII.17, United States, 2014. Emerg. Infect. Dis. 2015, 21, 1477–1479. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, Y.; Ishikawa, M.; Shimizu, T.; Komane, A.; Kasuo, S.; Shinohara, M.; Nagasawa, K.; Kimura, H.; Ryo, A.; Okabe, N.; et al. Genetic analyses of G2.17 norovirus strains in diarrheal disease outbreaks from December 2014 to March 2015 in Japan. Euro Surveill. 2015, 20, 21173. [Google Scholar] [CrossRef] [PubMed]


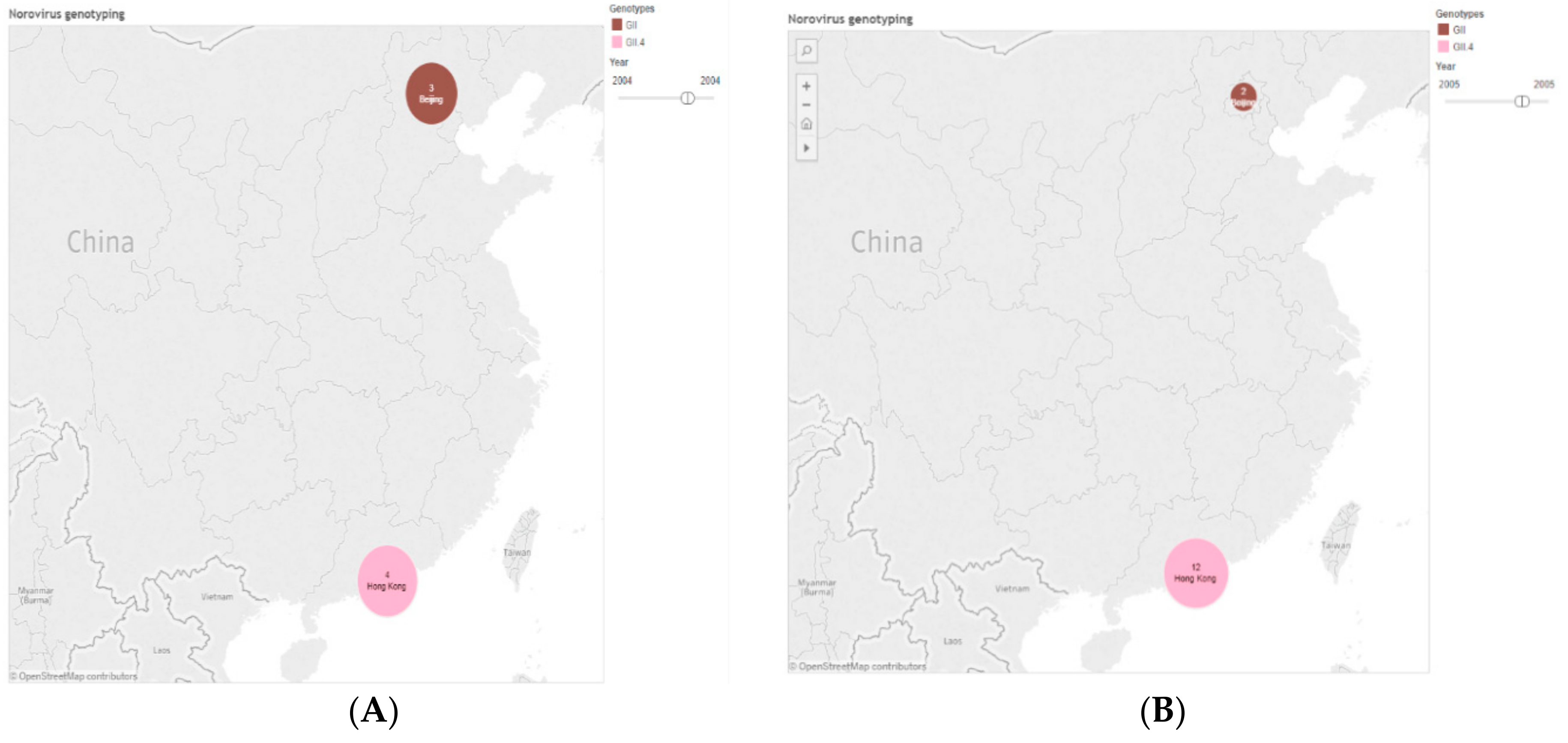
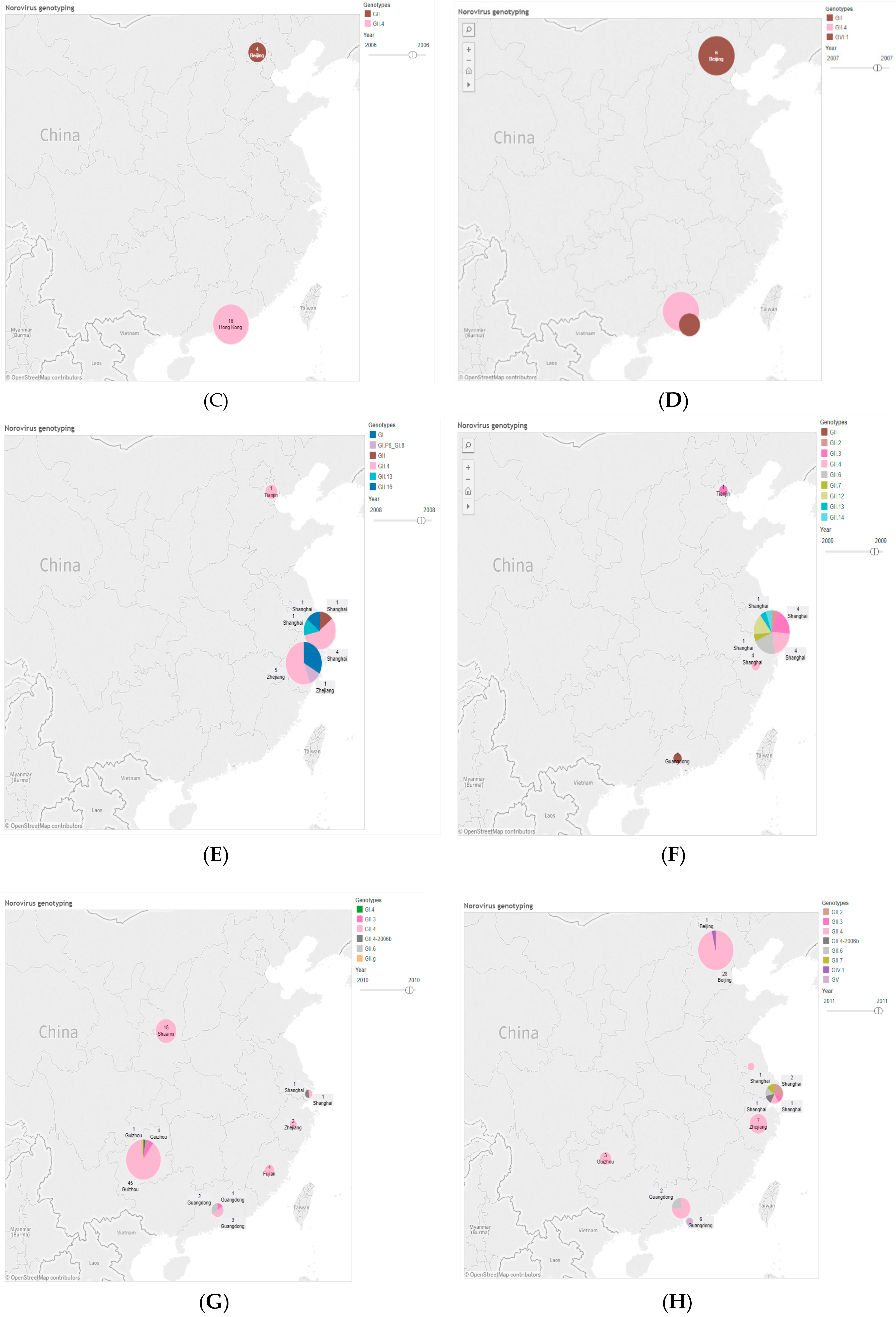
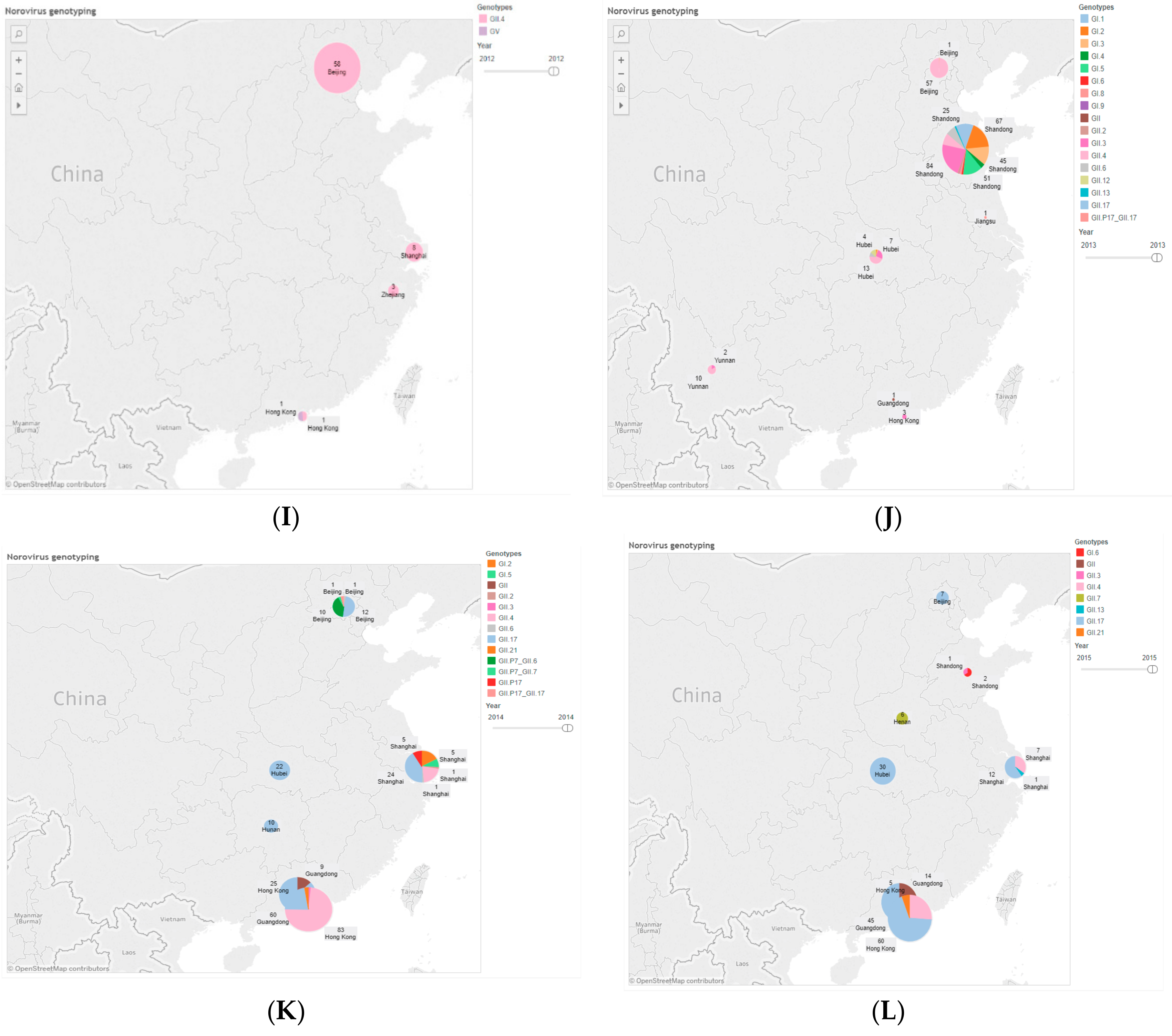
| Province | 2006 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | Total (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Anhui | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 1 | 4 (3.0) |
| Chongqing | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 4 (3.0) |
| Fujian | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 4 (3.0) |
| Guangdong | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 15 | 33 | 6 | 63 (47.7) |
| Guangxi | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 (0.8) |
| Hainan | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 (1.5) |
| Hebei | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 (0.8) |
| Hubei | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 4 (3.0) |
| Hunan | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 3 (2.3) |
| Jiangsu | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 9 | 2 | 17 (12.9) |
| Tianjin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 (0.8) |
| Zhejiang | 1 | 5 | 2 | 1 | 1 | 3 | 1 | 5 | 7 | 2 | 28 (21.2) |
| Total (%) | 1 (0.8) | 5 (3.8) | 2 (1.5) | 1 (0.8) | 1 (0.8) | 3 (2.3) | 13 (9.8) | 30 (22.7) | 61 (46.2) | 15 (11.3) | 132 (100.0) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, S.-W.; Chan, T.-C.; Cai, J.; Zhao, N.; Miao, Z.-P.; Chen, Y.-J.; Liu, S.-L. Genotypic and Epidemiological Trends of Acute Gastroenteritis Associated with Noroviruses in China from 2006 to 2016. Int. J. Environ. Res. Public Health 2017, 14, 1341. https://doi.org/10.3390/ijerph14111341
Qin S-W, Chan T-C, Cai J, Zhao N, Miao Z-P, Chen Y-J, Liu S-L. Genotypic and Epidemiological Trends of Acute Gastroenteritis Associated with Noroviruses in China from 2006 to 2016. International Journal of Environmental Research and Public Health. 2017; 14(11):1341. https://doi.org/10.3390/ijerph14111341
Chicago/Turabian StyleQin, Shu-Wen, Ta-Chien Chan, Jian Cai, Na Zhao, Zi-Ping Miao, Yi-Juan Chen, and She-Lan Liu. 2017. "Genotypic and Epidemiological Trends of Acute Gastroenteritis Associated with Noroviruses in China from 2006 to 2016" International Journal of Environmental Research and Public Health 14, no. 11: 1341. https://doi.org/10.3390/ijerph14111341






