Characterization of Carbonic Anhydrase 9 in the Alimentary Canal of Aedes aegypti and Its Relationship to Homologous Mosquito Carbonic Anhydrases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mosquito Rearing
2.2. Data Mining and Bioinformatics
2.3. Polymerase Chain Reaction (PCR)
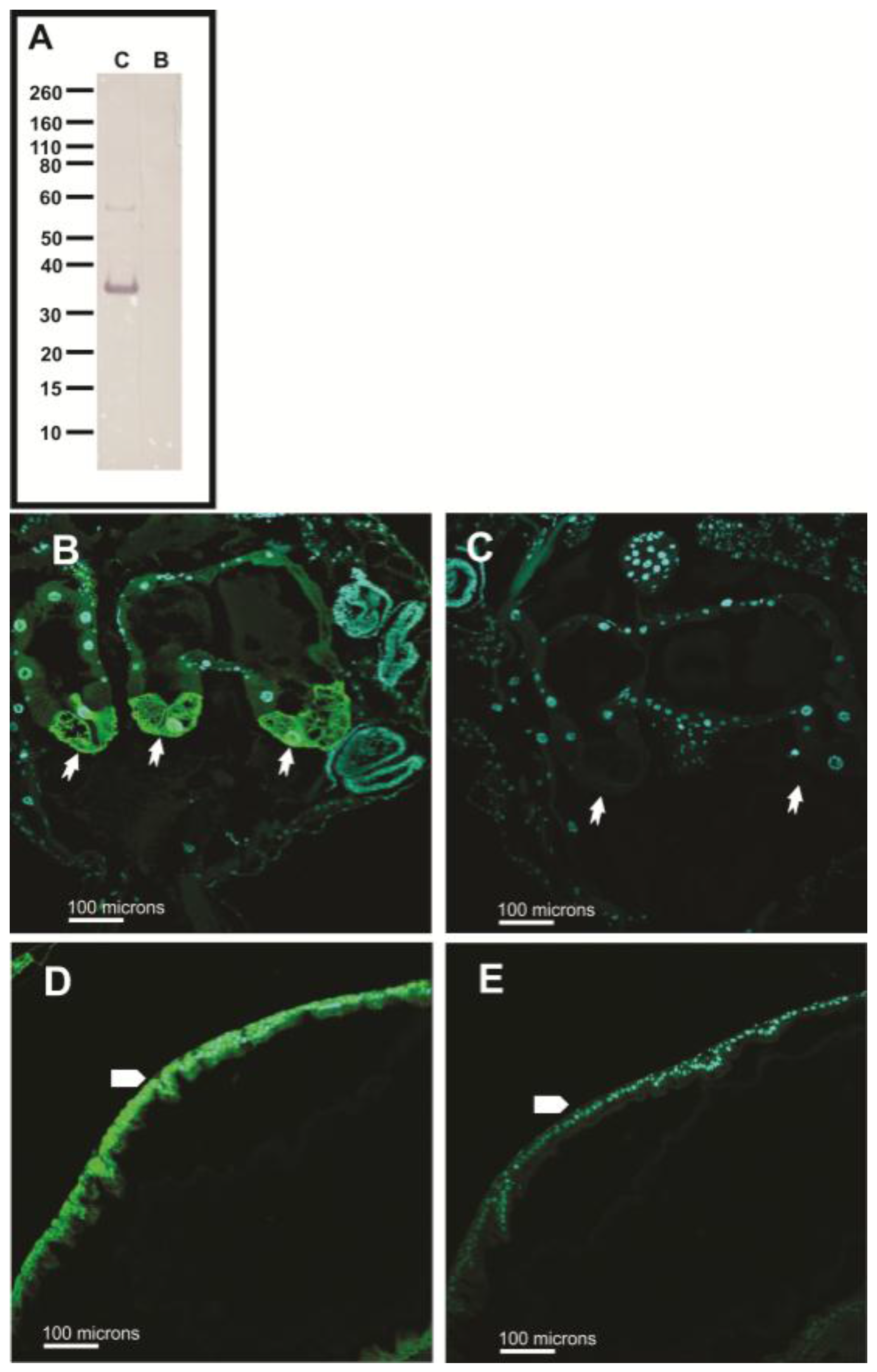
2.4. Western Blots
2.5. Immunohistochemical Analyses
2.6. Preabsorption Controls
3. Results
3.1. Predicted Transcript and Protein Sequence Information for Aedes aegypti CAs
3.1.1. AeCA Genome and Transcript Sequence Analysis
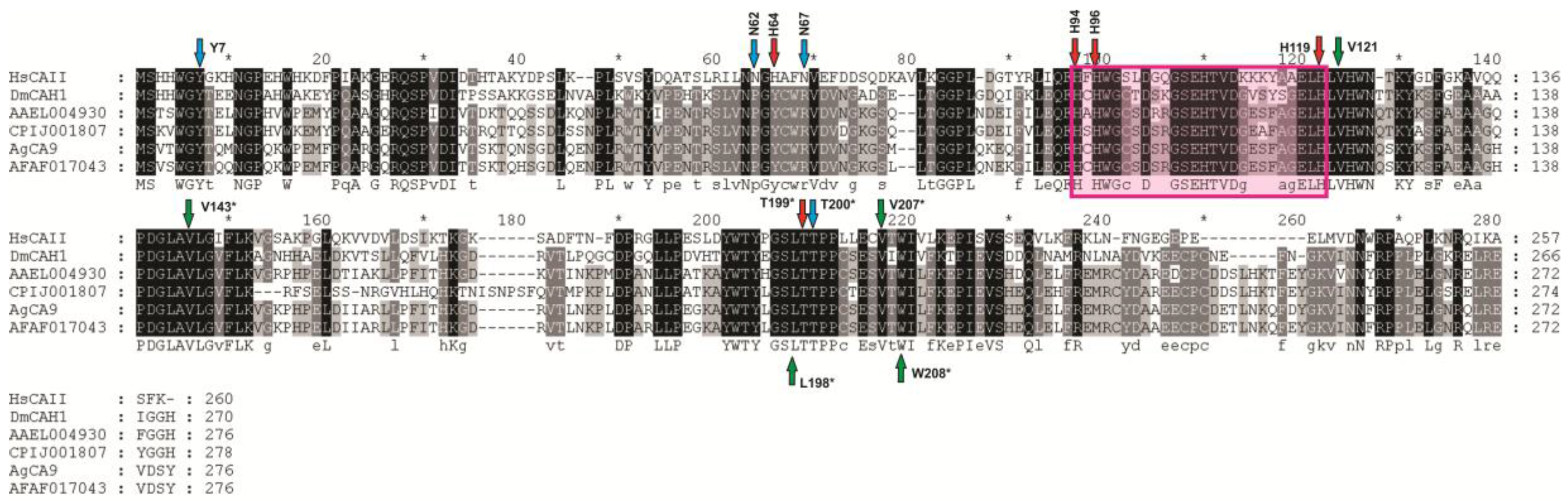
3.1.2. AeCA Protein Sequence Analysis
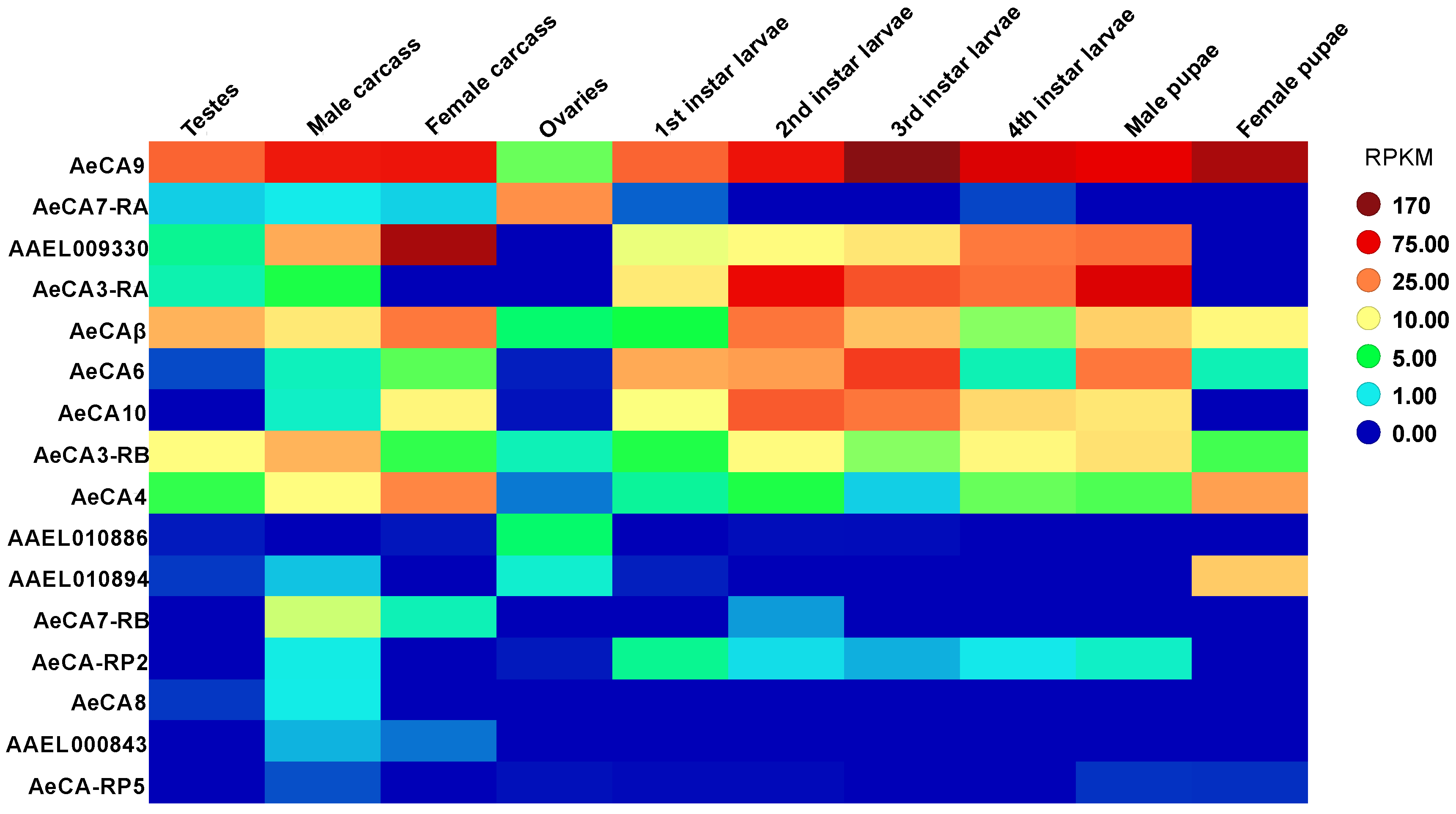
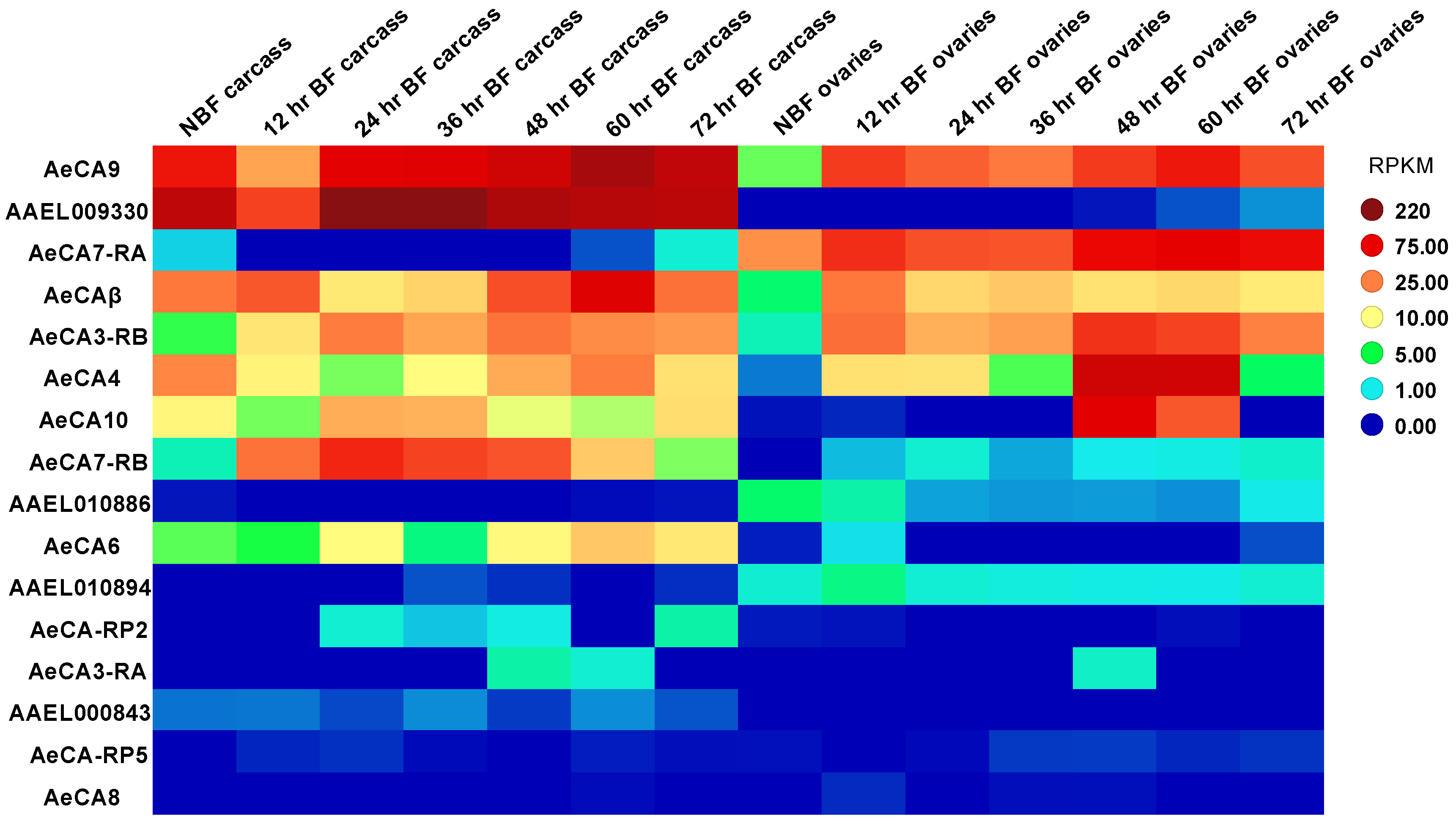
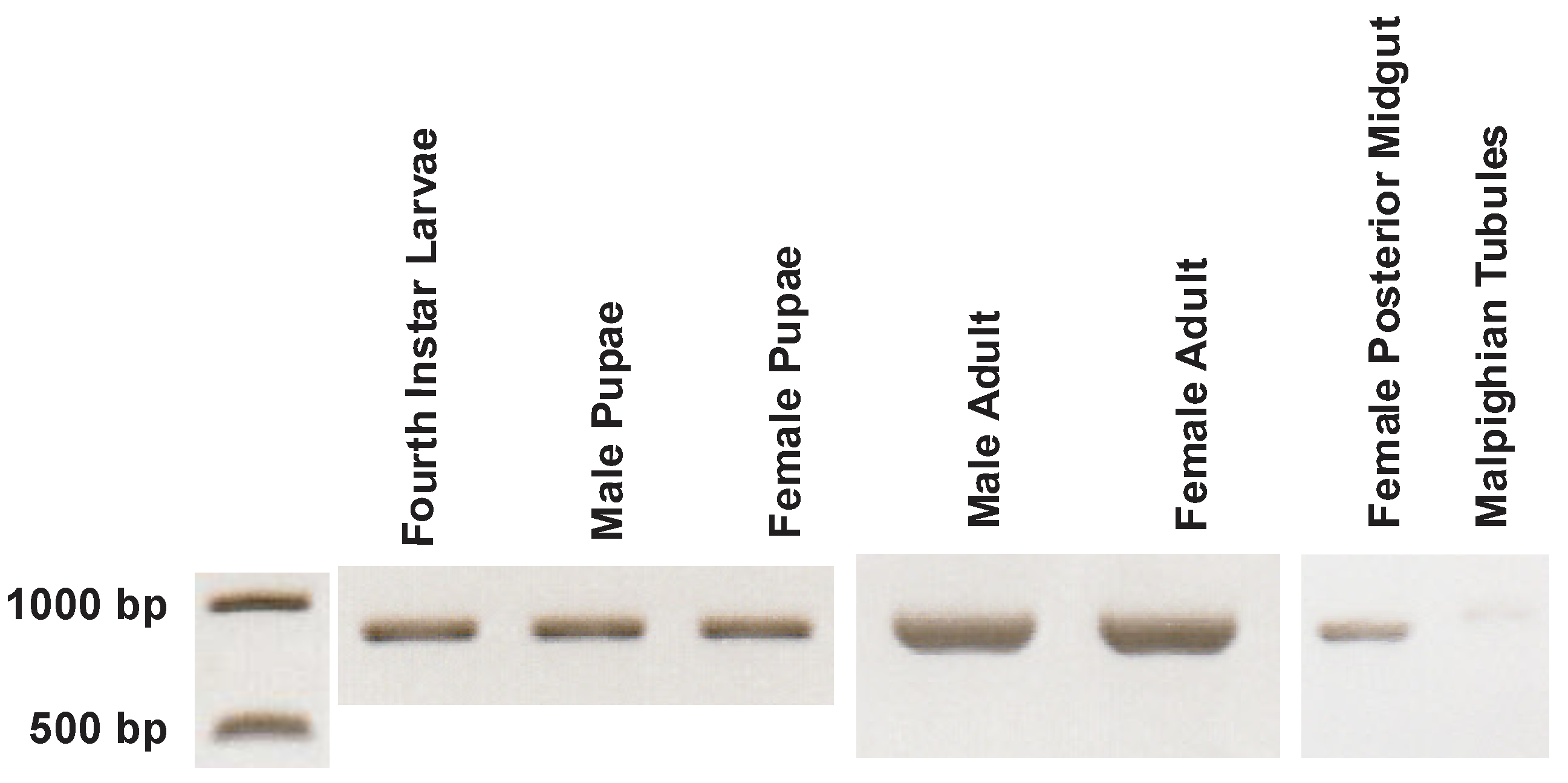
3.2. CA Transcript Expression across Development and Sex of Aedes aegypti
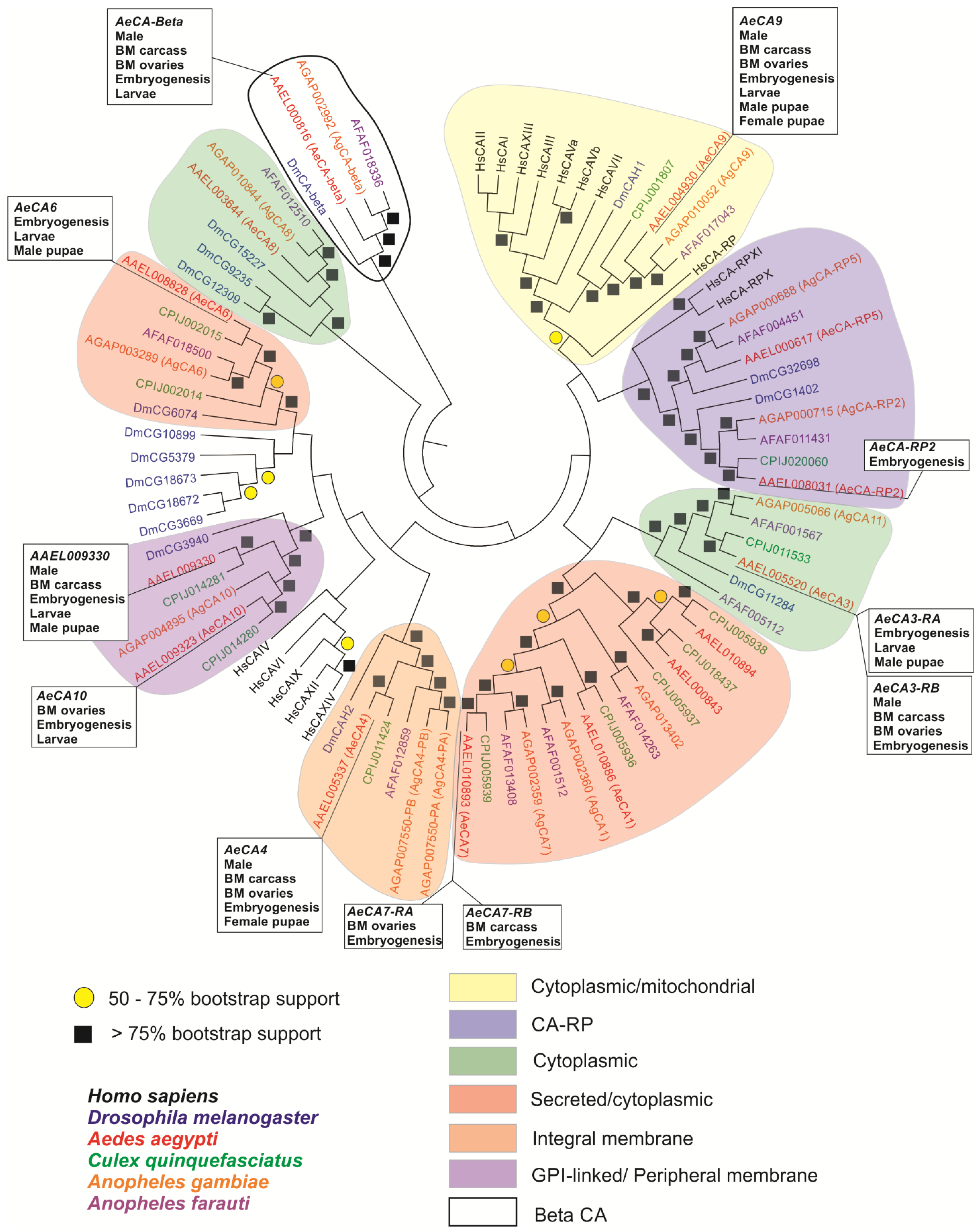
3.3. A Molecular Phylogeny of AeCA Protein Sequences
3.4. AeCA9 Transcript and Protein Expression across the Development and Sex of Aedes aegypti
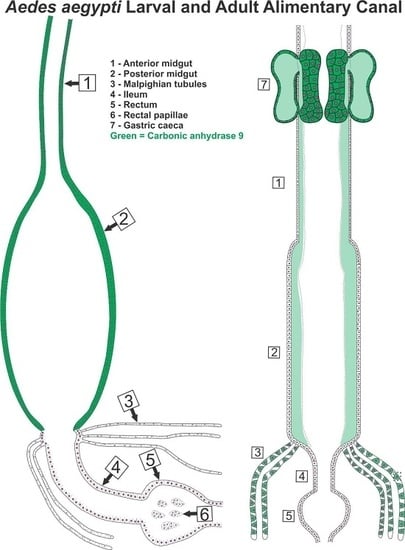
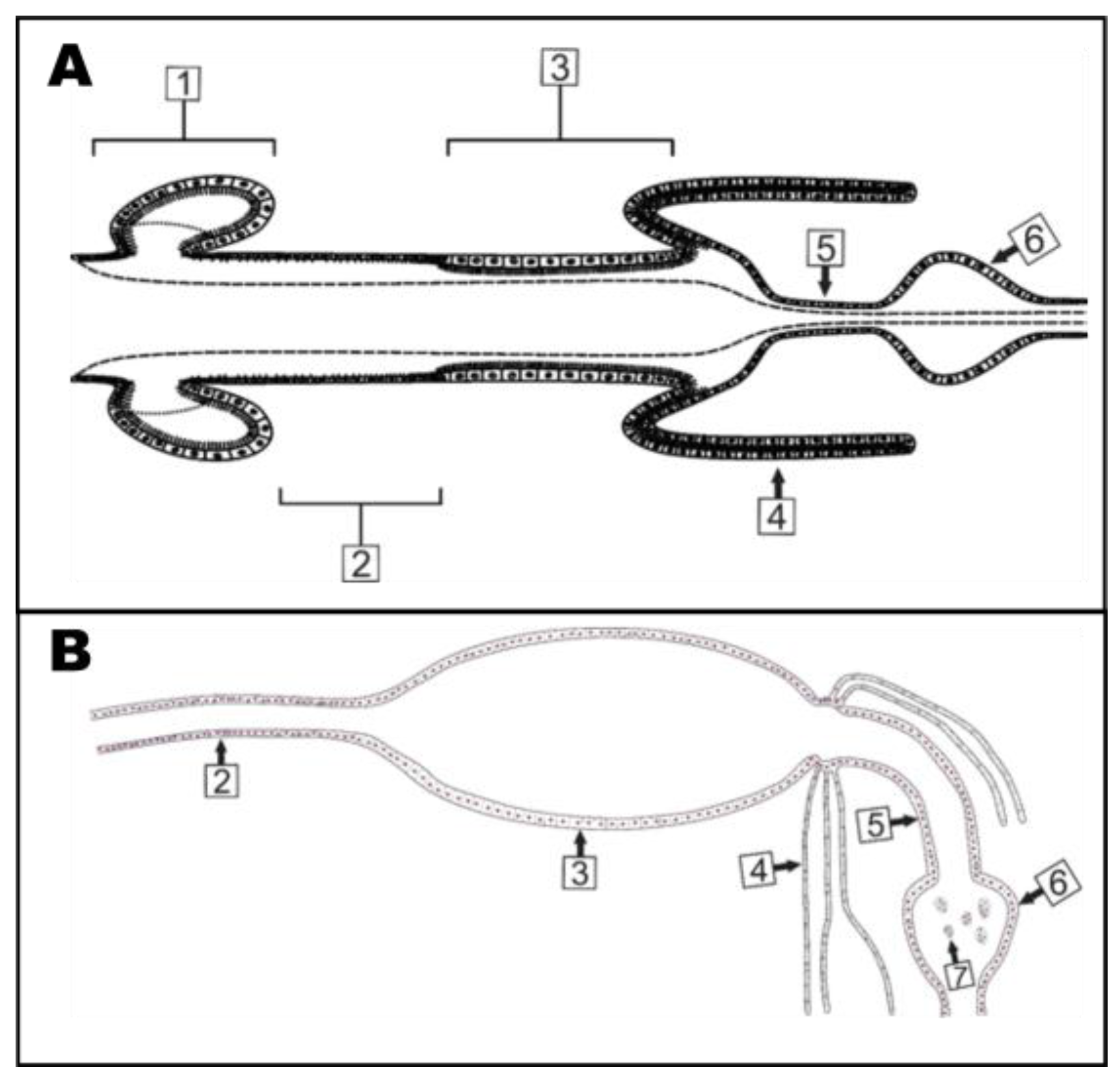
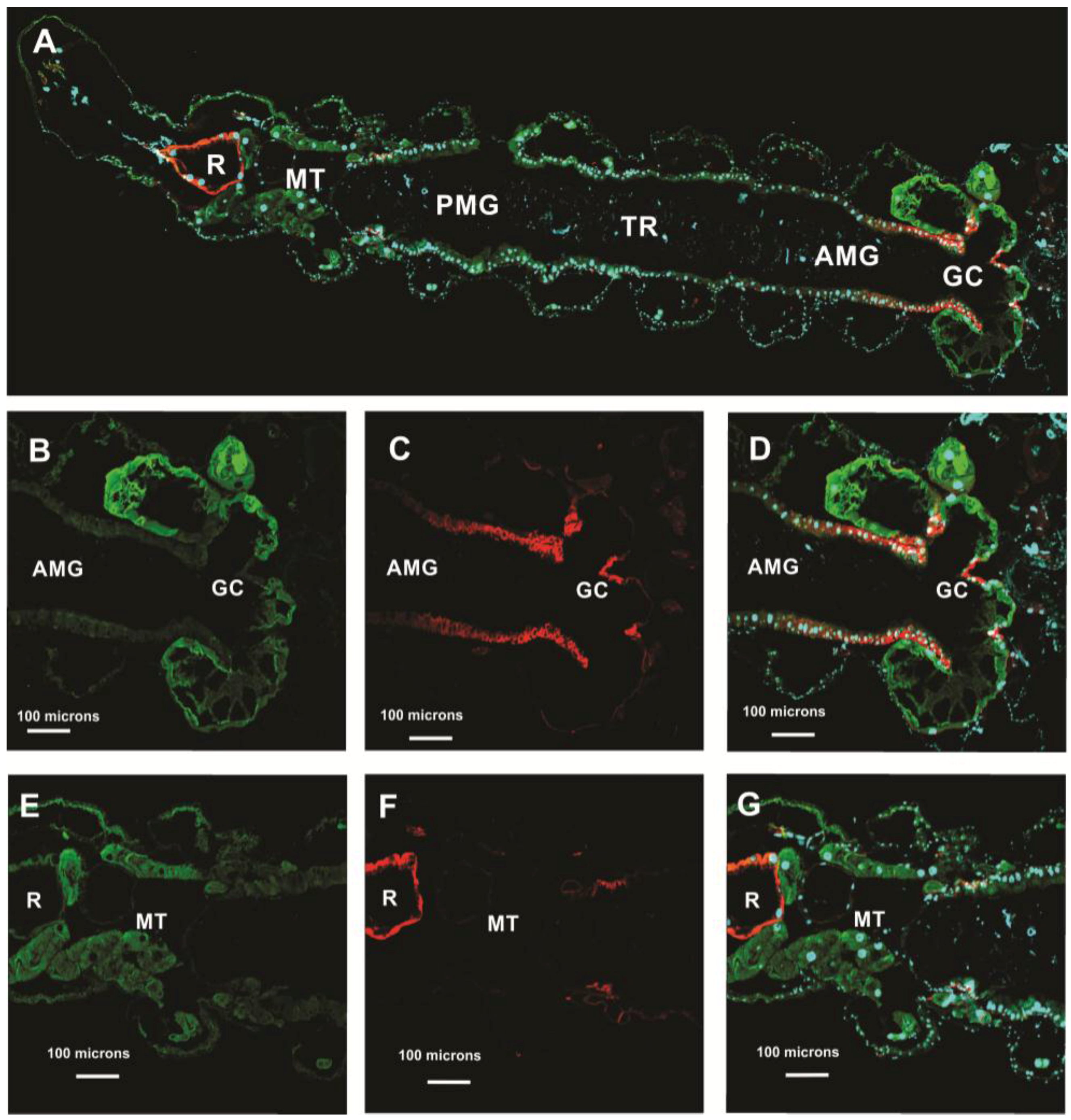
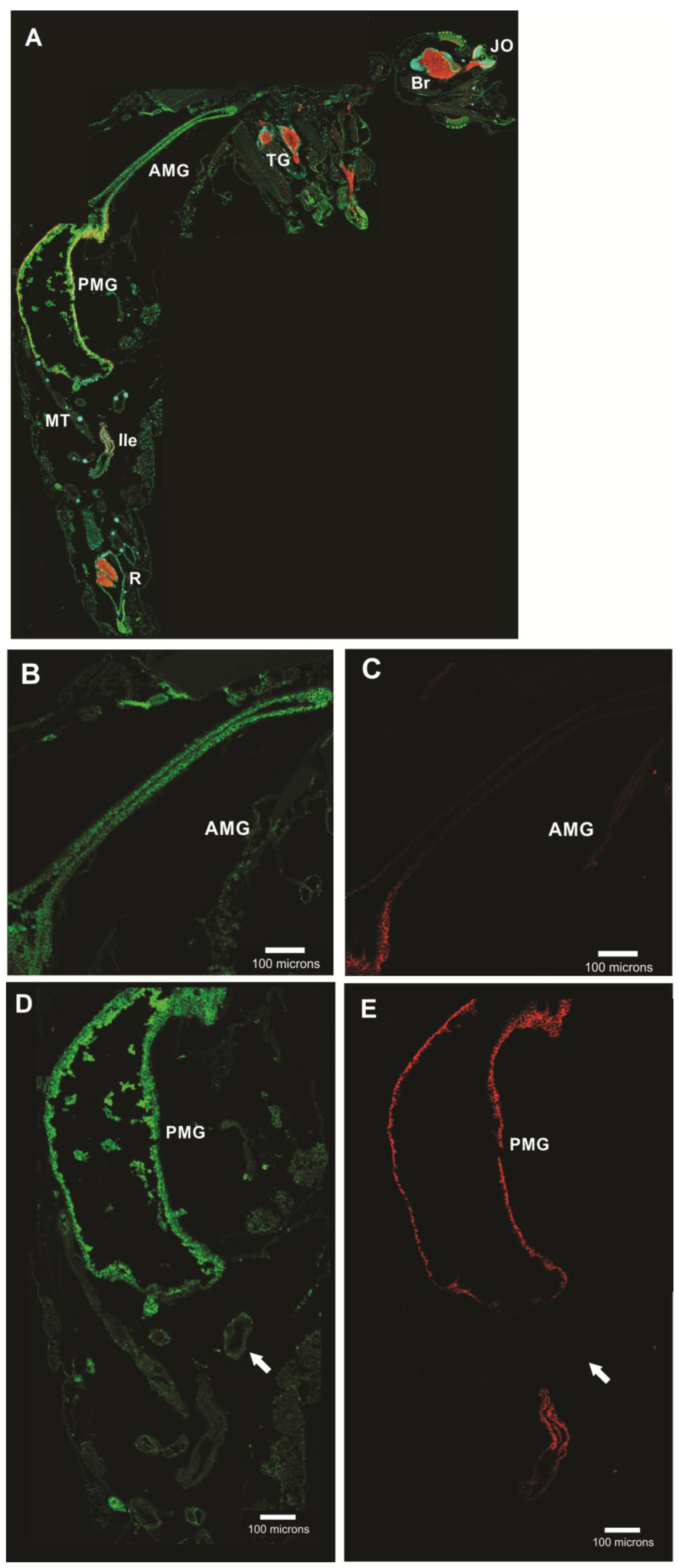
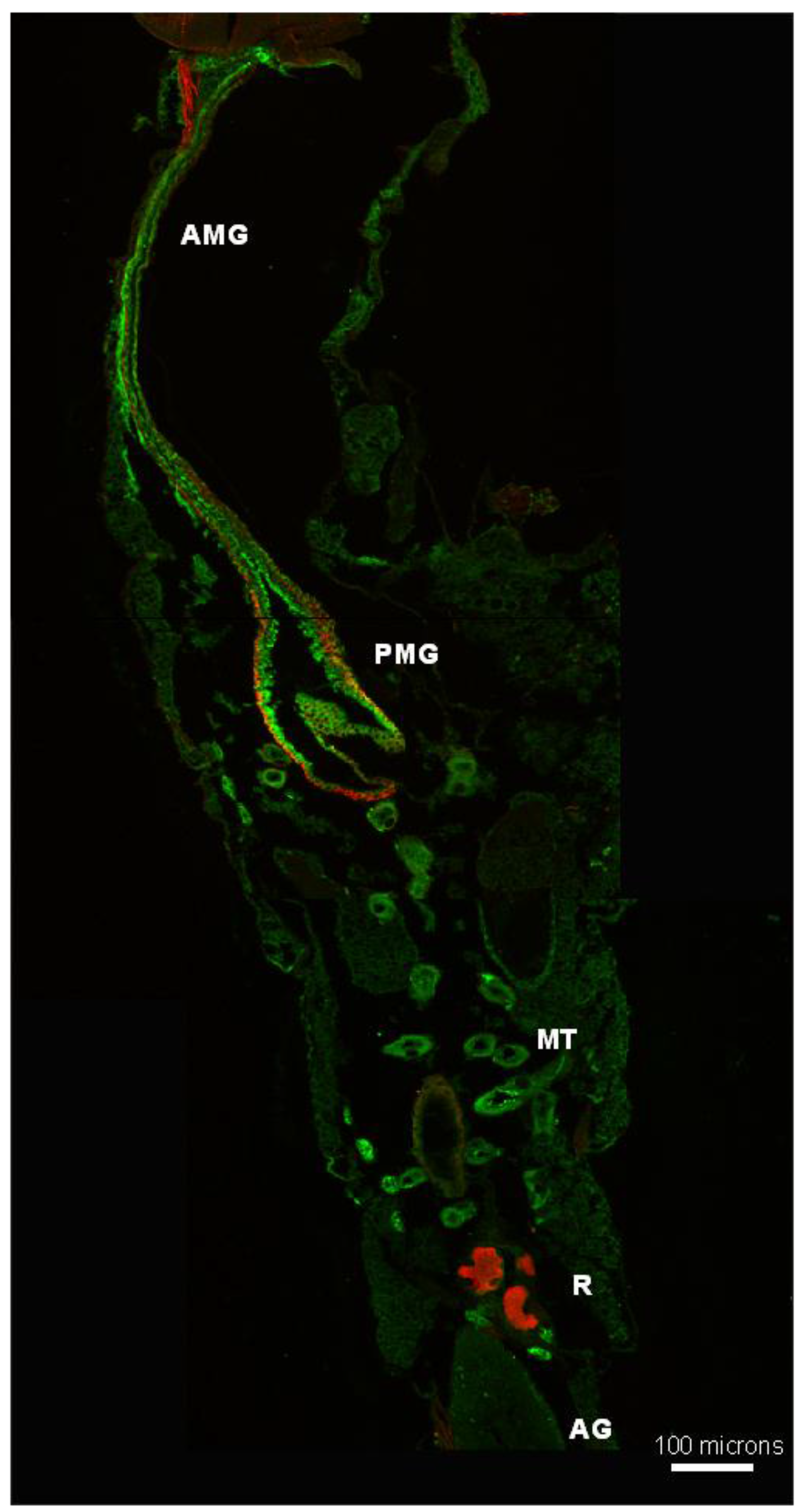
3.5. Subcellular Compartmentalization of AeCA9 in Aedes aegypti
3.5.1. Aedes aegypti Larvae
3.5.2. Aedes aegypti Adults
4. Discussion
4.1. AeCA9 Resembles the Most Ancestral Alpha CA and Is Widely Expressed in Aedes aegypti
4.2. AeCA9 Is Compartmentalized to the Alimentary Canal in Both Larvae and Adults
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Noriega, F.G.; Edgar, K.A.; Bechet, R.; Wells, M.A. Midgut exopeptidase activities in Aedes aegypti are induced by blood feeding. J. Insect Physiol. 2002, 48, 205–212. [Google Scholar] [CrossRef]
- Dadd, R.H. Alkalinity within the midgut of mosquito larvae with alkaline-active digestive enzymes. J. Insect Physiol. 1975, 21, 1847–1853. [Google Scholar] [CrossRef]
- Del Pilar Corena, M.; VanEkeris, L.; Salazar, M.I.; Bowers, D.; Fiedler, M.M.; Silverman, D.; Tu, C.; Linser, P.J. Carbonic anhydrase in the adult mosquito midgut. J. Exp. Biol. 2005, 208, 3263–3273. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.E.; VanEkeris, L.A.; Linser, P.J. Cloning and characterization of AgCA9, a novel alpha-carbonic anhydrase from Anopheles gambiae Giles sensu stricto (Diptera: Culicidae) larvae. J. Exp. Biol. 2007, 210, 3919–3930. [Google Scholar] [CrossRef] [PubMed]
- Linser, P.J.; Smith, K.E.; Seron, T.J.; Neira Oviedo, M. Carbonic anhydrases and anion transport in mosquito midgut pH regulation. J. Exp. Biol. 2009, 212, 1662–1671. [Google Scholar] [CrossRef] [PubMed]
- Akbari, O.S.; Antoshechkin, I.; Amrhein, H.; Williams, B.; Diloreto, R.; Sandler, J.; Hay, B.A. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 2013, 3, 1493–1509. [Google Scholar] [CrossRef] [PubMed]
- McKenna, R.; Frost, S.C. Overview of the carbonic anhydrase family. Subcell. Biochem. 2014, 75, 3–5. [Google Scholar] [PubMed]
- De Simone, G.; Di Fiore, A.; Capasso, C.; Supuran, C.T. The zinc coordination pattern in the η-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg. Med. Chem. Lett. 2015, 25, 1385–1389. [Google Scholar] [CrossRef] [PubMed]
- Hilvo, M.; Baranauskiene, L.; Salzano, A.M.; Scaloni, A.; Matulis, D.; Innocenti, A.; Scozzafava, A.; Monti, S.M.; Di Fiore, A.; De Simone, G.; et al. Biochemical characterization of CA IX, one of the most active carbonic anhydrase isozymes. J. Biol. Chem. 2008, 283, 27799–27809. [Google Scholar] [CrossRef] [PubMed]
- Lindskog, S. Structure and mechanism of carbonic anhydrase. Pharmacol. Ther. 1997, 74, 1–20. [Google Scholar] [CrossRef]
- Buren, S.; Ortega-Villasante, C.; Blanco-Rivero, A.; Martinez-Bernardini, A.; Shutova, T.; Shevela, D.; Messinger, J.; Bako, L.; Villarejo, A.; Samuelsson, G. Importance of post-translational modifications for functionality of a chloroplast-localized carbonic anhydrase (CAH1) in Arabidopsis thaliana. PLoS ONE 2011, 6, e21021. [Google Scholar] [CrossRef] [PubMed]
- Purkerson, J.M.; Schwartz, G.J. The role of carbonic anhydrases in renal physiology. Kidney Int. 2007, 71, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Postel, R.; Sonnenberg, A. Carbonic anhydrase 5 regulates acid-base homeostasis in zebrafish. PLoS ONE 2012, 7, e39881. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.P.; Swenson, E.R. The distribution and physiological significance of carbonic anhydrase in vertebrate gas exchange organs. Resp. Physiol. 2000, 121, 1–12. [Google Scholar] [CrossRef]
- Del Pilar Corena, M.; Fiedler, M.M.; Van Ekeris, L.; Tu, C.; Silverman, D.N.; Linser, P.J. Alkalization of larval mosquito midgut and the role of carbonic anhydrase in different species of mosquitoes. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2004, 137, 207–225. [Google Scholar] [CrossRef] [PubMed]
- Boudko, D.Y.; Moroz, L.L.; Harvey, W.R.; Linser, P.J. Alkalinization by chloride/bicarbonate pathway in larval mosquito midgut. Proc. Natl. Acad. Sci. USA 2001, 98, 15354–15359. [Google Scholar] [CrossRef] [PubMed]
- Onken, H.; Moffett, D.F. Fluid absorption in the isolated midgut of adult female yellow fever mosquitoes (Aedes aegypti). J. Exp. Biol. 2015, 218, 2023–2029. [Google Scholar] [CrossRef] [PubMed]
- Onken, H.; Moffett, S.B.; Moffett, D.F. Alkalinization in the isolated and perfused anterior midgut of the larval mosquito Aedes aegypti. J. Insect Sci. 2008, 8, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Linser, P.J.; Neira Oviedo, M.; Hirata, T.; Seron, T.J.; Smith, K.E.; Piermarini, P.M.; Romero, M.F. Slc4-like anion transporters of the larval mosquito alimentary canal. J. Insect Physiol. 2012, 58, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Clements, A.N. The Biology of Mosquitoes; Chapman and Hall: London, UK, 1992. [Google Scholar]
- Volkmann, A.; Peters, W. Investigations on the midgut caeca of mosquito larvae-II. Functional aspects. Tissue Cell 1989, 21, 253–261. [Google Scholar] [CrossRef]
- Moffett, S.B.; Moffett, D.F. Comparison of immunoreactivity to serotonin, FMRFamide and SCPb in the gut and visceral nervous system of larvae, pupae and adults of the yellow fever mosquito Aedes aegypti. J. Insect Sci. 2005, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Patrick, M.L.; Aimanova, K.; Sanders, H.R.; Gill, S.S. P-type Na+/K+-ATPase and V-type H+-ATPase expression patterns in the osmoregulatory organs of larval and adult mosquito Aedes aegypti. J. Exp. Biol. 2006, 209, 4638–4651. [Google Scholar] [CrossRef] [PubMed]
- Seron, T.J.; Hill, J.; Linser, P.J. A GPI-linked carbonic anhydrase expressed in the larval mosquito midgut. J. Exp. Biol. 2004, 207, 4559–4572. [Google Scholar] [CrossRef] [PubMed]
- Cocke, J.; Bridges, A.C.; Mayer, R.T.; Olson, J.K. Morphological effects of insect growth regulating compounds on Aedes aegypti (Diptera: Culicidae) larvae. Life Sci. 1979, 24, 817–831. [Google Scholar] [CrossRef]
- Corena, M.; Seron, T.J.; Lehman, H.K.; Ochrietor, J.D.; Kohn, A.; Tu, C.; Linser, P.J. Carbonic anhydrase in the midgut of larval Aedes aegypti: Cloning, localization and inhibition. J. Exp. Biol. 2002, 205, 591–602. [Google Scholar] [PubMed]
- Jagadeshwaran, U.; Onken, H.; Hardy, M.; Moffett, S.B.; Moffett, D.F. Cellular mechanisms of acid secretion in the posterior midgut of the larval mosquito (Aedes aegypti). J. Exp. Biol. 2010, 213, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Linser, P.J.; Harvey, W.R. Antibody to H+V-ATPase subunit E colocalizes with portasomes in alkaline larval midgut of freshwater mosquito (Aedes aegypti). J. Exp. Biol. 1999, 202, 2449–2460. [Google Scholar] [PubMed]
- Foster, W.A. Mosquito sugar feeding and reproductive energetics. Annu. Rev. Entomol. 1995, 40, 443–474. [Google Scholar] [CrossRef] [PubMed]
- Okuda, K.; Caroci, A.; Ribolla, P.; Marinotti, O.; de Bianchi, A.G.; Bijovsky, A.T. Morphological and enzymatic analysis of the midgut of Anopheles darlingi during blood digestion. J. Insect Physiol. 2005, 51, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Pullikuth, A.K.; Aimanova, K.; Kangethe, W.; Sanders, H.R.; Gill, S.S. Molecular characterization of sodium/proton exchanger 3 (NHE3) from the yellow fever vector, Aedes aegypti. J. Exp. Biol. 2006, 209, 3529–3544. [Google Scholar] [CrossRef] [PubMed]
- Pacey, E.K.; O′Donnell, M.J. Transport of H(+), Na(+) and K(+) across the posterior midgut of blood-fed mosquitoes (Aedes aegypti). J. Insect Physiol. 2014, 61, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Piermarini, P.M.; Grogan, L.F.; Lau, K.; Wang, L.; Beyenbach, K.W. A SLC4-like anion exchanger from renal tubules of the mosquito (Aedes aegypti): Evidence for a novel role of stellate cells in diuretic fluid secretion. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 298, R642–R660. [Google Scholar] [CrossRef] [PubMed]
- Giraldo-Calderón, G.I.; Emrich, S.J.; MacCallum, R.M.; Maslen, G.; Dialynas, E.; Topalis, P.; Ho, N.; Gesing, S.; Consortium, T.V.; Madey, G.; et al. VectorBase: An updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015, 43, D707–D713. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.; Chang, H.Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef] [PubMed]
- Kall, L.; Krogh, A.; Sonnhammer, E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.E.; VanEkeris, L.A.; Okech, B.A.; Harvey, W.R.; Linser, P.J. Larval anopheline mosquito recta exhibit a dramatic change in localization patterns of ion transport proteins in response to shifting salinity: A comparison between anopheline and culicine larvae. J. Exp. Biol. 2008, 211, 3067–3076. [Google Scholar] [CrossRef] [PubMed]
- Lebovitz, R.M.; Takeyasu, K.; Fambrough, D.M. Molecular characterization and expression of the (Na+ + K+)-ATPase alpha-subunit in Drosophila melanogaster. EMBO J. 1989, 8, 193–202. [Google Scholar] [PubMed]
- Juneja, P.; Osei-Poku, J.; Ho, Y.S.; Ariani, C.V.; Palmer, W.J.; Pain, A.; Jiggins, F.M. Assembly of the genome of the disease vector Aedes aegypti onto a Genetic linkage map allows mapping of genes affecting disease transmission. PLoS Negl. Trop. Dis. 2014, 8, e2652. [Google Scholar] [CrossRef] [PubMed]
- Eisenhaber, B.; Bork, P.; Eisenhaber, F. Prediction of potential GPI-modification sites in proprotein sequences. J. Mol. Biol. 1999, 292, 741–758. [Google Scholar] [CrossRef] [PubMed]
- Fisher, S.Z.; Tariku, I.; Case, N.M.; Tu, C.; Seron, T.; Silverman, D.N.; Linser, P.J.; McKenna, R. Expression, purification, kinetic, and structural characterization of an alpha-class carbonic anhydrase from Aedes aegypti (AaCA1). Biocim. Biophys. Acta 2006, 1764, 1413–1419. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, N.; Jackson, D.; Marie, B.; Ramos-Silva, P.; Marin, F. The evolution of metazoan α-carbonic anhydrases and their roles in calcium carbonate biomineralization. Front. Zool. 2014, 11, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Wessing, A.; Zierold, K.; Bertram, G. Carbonic anhydrase supports electrolyte transport in Drosophila Malpighian tubules. Evidence by X-ray microanalysis of cryosections. J. Insect Physiol. 1997, 43, 17–28. [Google Scholar] [CrossRef]
- Palatroni, P.; Gabrielli, M.G.; Scattolini, B. Histochemical localization of carbonic anhydrase in Malpighian tubules of Culex pipiens. Experientia 1981, 37, 409–411. [Google Scholar] [CrossRef]
- Evans, A.M.; Aimanova, K.G.; Gill, S.S. Characterization of a blood-meal-responsive proton-dependent amino acid transporter in the disease vector, Aedes aegypti. J. Exp. Biol. 2009, 212, 3263–3271. [Google Scholar] [CrossRef] [PubMed]

| Isoform | Genome Sequence Length | Transcript Sequence Length | 5′ UTR Length | 3′ UTR Length | Coding Sequence Length | Amino Acids |
|---|---|---|---|---|---|---|
| AAEL000617 | 6274 | 1452 | 0 | 0 | 1452 | 483 |
| AAEL000816 | 1015 | 948 | 36 | 261 | 651 | 216 |
| AAEL000843 | 877 | 822 | 0 | 0 | 822 | 273 |
| AAEL003644 | 986 | 986 | 0 | 0 | 987 | 328 |
| AAEL004930 | 54,321 | 2742 | 801 | 1110 | 831 | 276 |
| AAEL005337 | 32,439 | 1530 | 0 | 486 | 1044 | 347 |
| AAEL005520-RA | 16,994 | 1082 | 147 | 164 | 771 | 256 |
| AAEL005520-RB | 16,994 | 1567 | 632 | 164 | 771 | 256 |
| AAEL008031 | 68,384 | 999 | 0 | 0 | 999 | 332 |
| AAEL008828 | 34,026 | 1737 | 444 | 348 | 945 | 314 |
| AAEL009323 | 28,262 | 1276 | 0 | 382 | 894 | 297 |
| AAEL009330 | 1157 | 1103 | 102 | 53 | 948 | 315 |
| AAEL010886 | 17,765 | 1657 | 509 | 125 | 1023 | 340 |
| AAEL010893-RA | 34,039 | 1773 | 253 | 380 | 1140 | 379 |
| AAEL010893-RB | 34,039 | 2215 | 695 | 380 | 1140 | 379 |
| AAEL010894 | 4503 | 1522 | 562 | 105 | 855 | 284 |
| Isoform | Signal Peptide | Transmembrane Helix | Predicted Localization | Predicted Orthology | Aegypti Identifiers |
|---|---|---|---|---|---|
| AAEL000617 | No | No | Cytoplasmic | AgCA-RP5 | AeCA-RP5 |
| AAEL000816 | No | No | Cytoplasmic | AgCAβ | AeCAβ |
| AAEL000843 | No | No | Cytoplasmic | AGAP013402 | AAEL000843 |
| AAEL003644 | No | No | Cytoplasmic | AgCA8 | AeCA8 |
| AAEL004930 | No | No | Cytoplasmic | AgCA9 | AeCA9 |
| AAEL005337 | No | Yes | Integral membrane | AgCA4 | AeCA4 |
| AAEL005520 | No | No | Mitochondrial | AgCA3/11 | AeCA3/11 |
| AAEL008031 | No | No | Cytoplasmic | AgCA-RP2 | AeCA-RP2 |
| AAEL008828 | Yes | No | Secreted | AgCA6 | AeCA6 |
| AAEL009323 | Yes | Yes | GPI-anchored | AgCA10 | AeCA10 |
| AAEL009330 | Yes | Yes | GPI-anchored | N/A | AAEL009330 |
| AAEL010886 | Yes | No | Secreted | AGAP002360 | AAEL010886 |
| AAEL010893 | Yes | No | Secreted | AgCA7 | AeCA7 |
| AAEL010894 | Yes | No | Secreted | AGAP013402 | AAEL010894 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dixon, D.P.; Van Ekeris, L.; Linser, P.J. Characterization of Carbonic Anhydrase 9 in the Alimentary Canal of Aedes aegypti and Its Relationship to Homologous Mosquito Carbonic Anhydrases. Int. J. Environ. Res. Public Health 2017, 14, 213. https://doi.org/10.3390/ijerph14020213
Dixon DP, Van Ekeris L, Linser PJ. Characterization of Carbonic Anhydrase 9 in the Alimentary Canal of Aedes aegypti and Its Relationship to Homologous Mosquito Carbonic Anhydrases. International Journal of Environmental Research and Public Health. 2017; 14(2):213. https://doi.org/10.3390/ijerph14020213
Chicago/Turabian StyleDixon, Daniel P., Leslie Van Ekeris, and Paul J. Linser. 2017. "Characterization of Carbonic Anhydrase 9 in the Alimentary Canal of Aedes aegypti and Its Relationship to Homologous Mosquito Carbonic Anhydrases" International Journal of Environmental Research and Public Health 14, no. 2: 213. https://doi.org/10.3390/ijerph14020213