1. Introduction
Antibiotic resistance represents a significant and complex global health problem. Global consumption of antibiotics has increased by nearly 40% in the last decade [
1]. Apart from fundamental applications in clinical settings, very large amounts of antibiotics are used in agriculture, the food industry, and aquaculture [
2]. Due to incomplete metabolism and the environmental spread of unused antibiotics, they enter the ecosystem, serving as a potent stimulus to elicit a bacterial adaptation response to develop antibiotic resistance and genes [
3,
4]. The accumulation of antibiotics in the environment facilitates the spread of antibiotic resistance genes. Various resistance mechanisms are continuously emerging and spreading globally, which threatens our ability to treat common infectious diseases, resulting in increased death, disability, and costs. TheWorld Health Assembly, in 2015, thus adopted a global action plan on antimicrobial resistance focussing on bacterial resistance [
5].
There is a worldwide concern about the emergence of antibiotic resistance in bacteria carried by healthy individuals, so-called commensal bacteria. Commensal bacteria from the gut microbes, e.g., coliforms, may play a crucial role in the spread of resistance within a community. Surveillance data shows that resistance in
Escherichia coli is generally consistently highest for antimicrobial agents that have been in use the longest time in human and veterinary medicine [
6].
E. coli is also considered an indicator bacteria of antibiotic resistance. Animal and human fecal flora and the environment, including water sources, serve as natural habitats and reservoirs of antibiotic-resistant bacteria and resistance genes. Antibiotic resistance in wastewater, surface water, and drinking water is well documented [
7,
8].
Thus, within the community, resistant bacteria circulated from person to person or from animals and environment to person, or vice versa. The epidemiology of antibiotic-resistant microorganisms at the human-animal-environmental interface involves complex and largely unpredictable systems that include transmission routes of resistant bacteria, as well as resistance genes and the impact of antibiotic-selective pressures in various reservoirs (animals, humans, and the environment). Though the presence and patterns of antibiotic resistant commensal indicator bacteria
E. coli isolates from humans, animals, and water have been studied in isolation, it is now recognized that they need to be studied together, i.e., using the ‘one-health’ approach [
5]. Thus, our aim is to determine and compare the antibiotic resistance pattern among commensal coliforms and
E. coli from humans, animals, and water from the same community.
4. Discussion
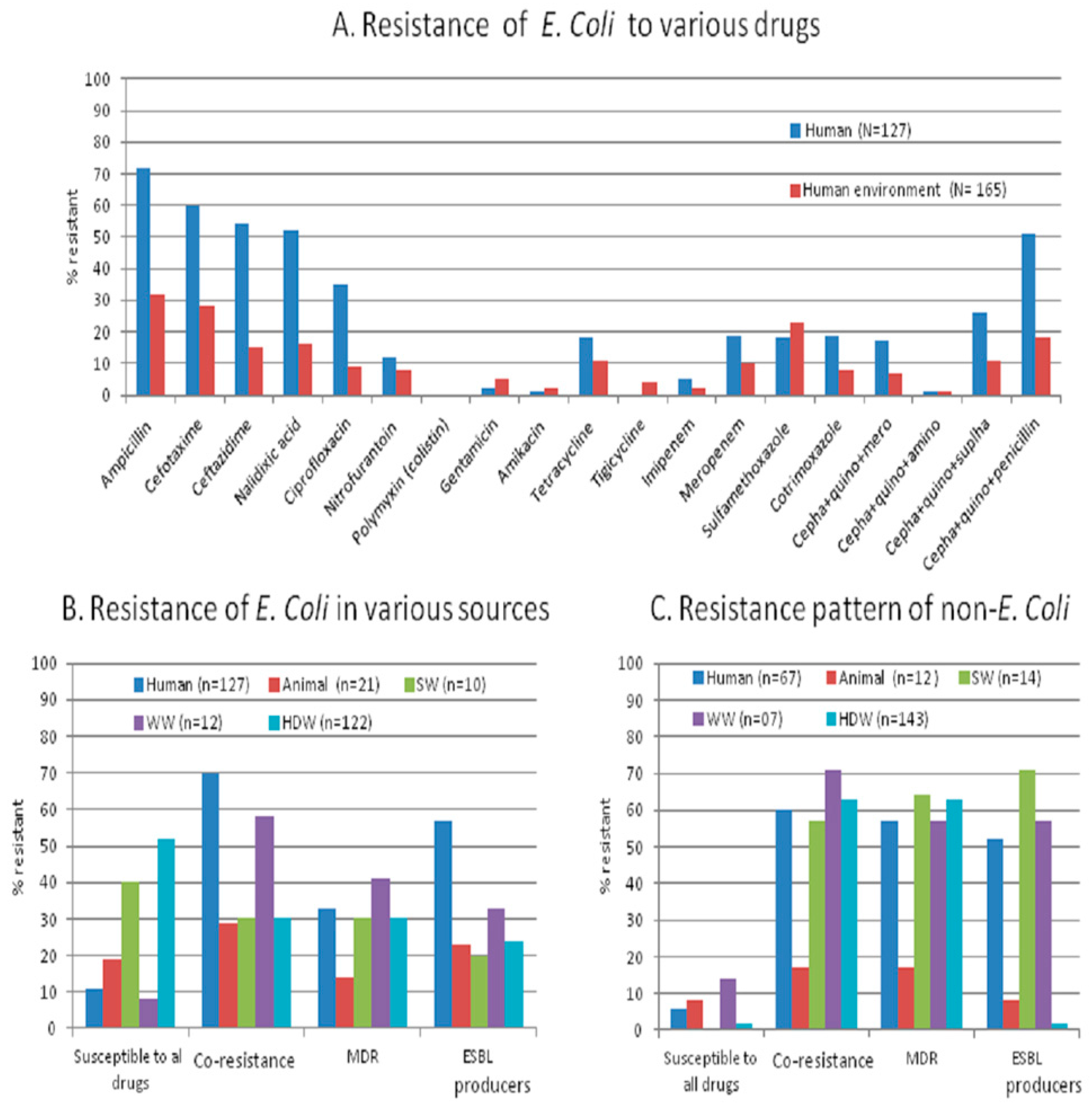
We studied antibiotic resistance and selected antibiotic resistance genes in human stool together with their shared and neighboring environment in a rural community from Central India with a ‘one-health’ approach. We found that the antibiotic resistance pattern and its genetic make-up are essentially the same in commensal bacteria from humans and their environment. The percentage of resistant isolates, including MDR (
Figure 1A,B), is higher in humans than in the environment (animal stool and water samples), but the load (number of resistant isolates/sample) is higher in the environment than in humans. The appearance of antibiotic-resistant bacteria in healthy individuals and their environment should be evaluated together to accomplish effective antibiotic resistance control.
The antibiotic resistance profile including certain patterns of co-resistance and MDR (i.e., cephalosporin-quinolone-penicillin, sulphonamide + tetracycline + cephalosporin, quinolones + carbapenem + sulfonamide or + tetracycline) in
E. coli obtained from humans, animals, source- and household-drinking water are high (57%–69%) in our study area. The presence of co-resistance and MDR signifies that there might be high use of antibiotics inhuman and non-human use in the community. The non-human use of highly-important antibiotics contributes to the resistance against a range of antibiotics [
1,
2,
15]. Van den Bogaard et al. and others have shown that the selective pressure on the commensal microflora due to antibiotic misuse determine the frequency and pattern of resistance in a population [
16]. The relatively cheap and commonly prescribed drugs commonly favour high co-resistance [
17,
18].
We found similar patterns of co-resistance, MDR, and gene carriage in various sources. Nearly 90% of MDR
E. coli isolates are carrying plasmid-encoded (
blaCTX-M1,
blaCTX-M9,
qnrS, and
qnrS) genes, which may indicate the possible spread of the resistance genes between diverse sources. This is similar to another study from India [
19].
CTX-M–producing
E. coli is the dominant MDR
E. coli in all parts of Asia and of major clinical significance [
20]. The patterns of antibiotic use in the community favor the persistence of plasmids carrying antibiotic resistance genes. The intestine is considered as a 'hot spot' for the transfer of resistance genes between bacteria as the exposure of frequently-used antibiotics to a high density of bacteria favours evolution and dissemination of antibiotic resistance by cell-to-cell contact [
21,
22]. Additionally, the existing various species of MDR bacteria, as we noticed in MDR non-
E. coli coliform species, (
Table 6,
Figure 1C), might also be contributing to the spread of antibiotic resistance genes in the intestine with
E. coli.
The resistant isolates are distributed in a higher percentage of drinking-water samples compared to human samples. In rural communities, the high level of bacterial contamination is reported in source-water to the extent that it lacks the criteria of safe-water supply for domestic purposes [
23]. Studies illustrate that surface water contamination occurs mainly from livestock operations and human sewage and that decreasing livestock access to surface water reduced the fecal coliforms levels by an average of 94% [
24]. Treatment processes of water, however, might further result in a selective increase of antibiotic-resistant bacteria and might, therefore, increase the occurrence of multidrug-resistant organisms [
11,
25]. It has also been observed that the microbiological quality of water in vessels in households is lower than that at the source, suggesting that bacterial contamination is widespread during collection, transport, storage, and drawing of water [
26].
In our study, phylogenetic group D (extra-intestinal virulent)
E. coli isolates with resistant genes are more often found from human stool than from environmental samples (30%–52% vs. 0%–24%). It has been reported that co-location of genes in plasmids not only results in resistance to multiple antibiotics, but also in the increased presence of virulence determinants, which facilitates infections [
27]. Indeed, the exposure of commensal bacteria to antibiotics increases the carriage level of resistant organisms that might result in the transmission of resistance to a virulent organism [
28]. Johnson et al. [
29] reported the horizontal transfer of antibiotic resistance not only between isolates from one source to another, but also from resistant to susceptible isolates in the same source. The number of virulent strains carrying resistant genes in human commensal samples is a matter of public health concern, as it may give rise to infection with an increased risk of treatment failure.
We have not identified any
E. coli or non-
E. coli isolates (including all forms of MDR strains) with colistin resistance or
mcr-1 gene carriage. With the emergence of MDR and extensive drug resistant (XDR) strains of Gram-negative bacteria, colistin is considered as one of the few last resort antibacterial agents. Recently, sporadic clinical cases infected with colistin-resistant
E. coli carrying the
mcr-1 gene has been described in India [
30,
31]. The plasmid-mediated
mcr-1 gene to colistin resistance is a matter of global alarm as its spread within the human commensal flora could lead to epidemics of virtually untreatable infections. Measures with the ‘one-health’ approach, such as colistin susceptibility testing of MDR isolates from patients, testing of food, animal, environmental isolates, and the reduction of colistin use in food-producing animals would be crucial for effective minimization of
mcr-
1-positive commensal dissemination in the community and healthcare facilities.
Our study has some methodological limitations. The study, being from a village, cannot be generalized. There is no reason, however, to believe that the situation in this village is very different from many other villages with similar low socio-economic levels in India. Additionally, in our study, none of the carbapenem-resistant isolates (six imipenem resistant and 29 meropenem resistant isolates) from all sources are carrying any of the tested (
NDM-1,
VIM, and
IMP) carbapenemases encoding genes. Studies showed the presence of
OXA-48 and
NDM-1 genes in clinical isolates from India [
32,
33]. However, in another study from our setting, we did not find any of these genes in either clinical or in hospital waste water [
34]. We, however, cannot rule out some different resistance mechanisms in these isolates, which we have not tested. Although our study involves a limited number of animals and sewage water samples, the comparison of multiple types of environmental samples with apparently healthy human samples from community provides us better understanding about the current scenario of antibiotic resistance at the community level. This is required in scientific research for establishing effective measures to mitigate resistance in clinically relevant bacteria.