Viability of Legionella pneumophila in Water Samples: A Comparison of Propidium Monoazide (PMA) Treatment on Membrane Filters and in Liquid
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Cultivation
2.2. Cytotoxic Effect of PMA
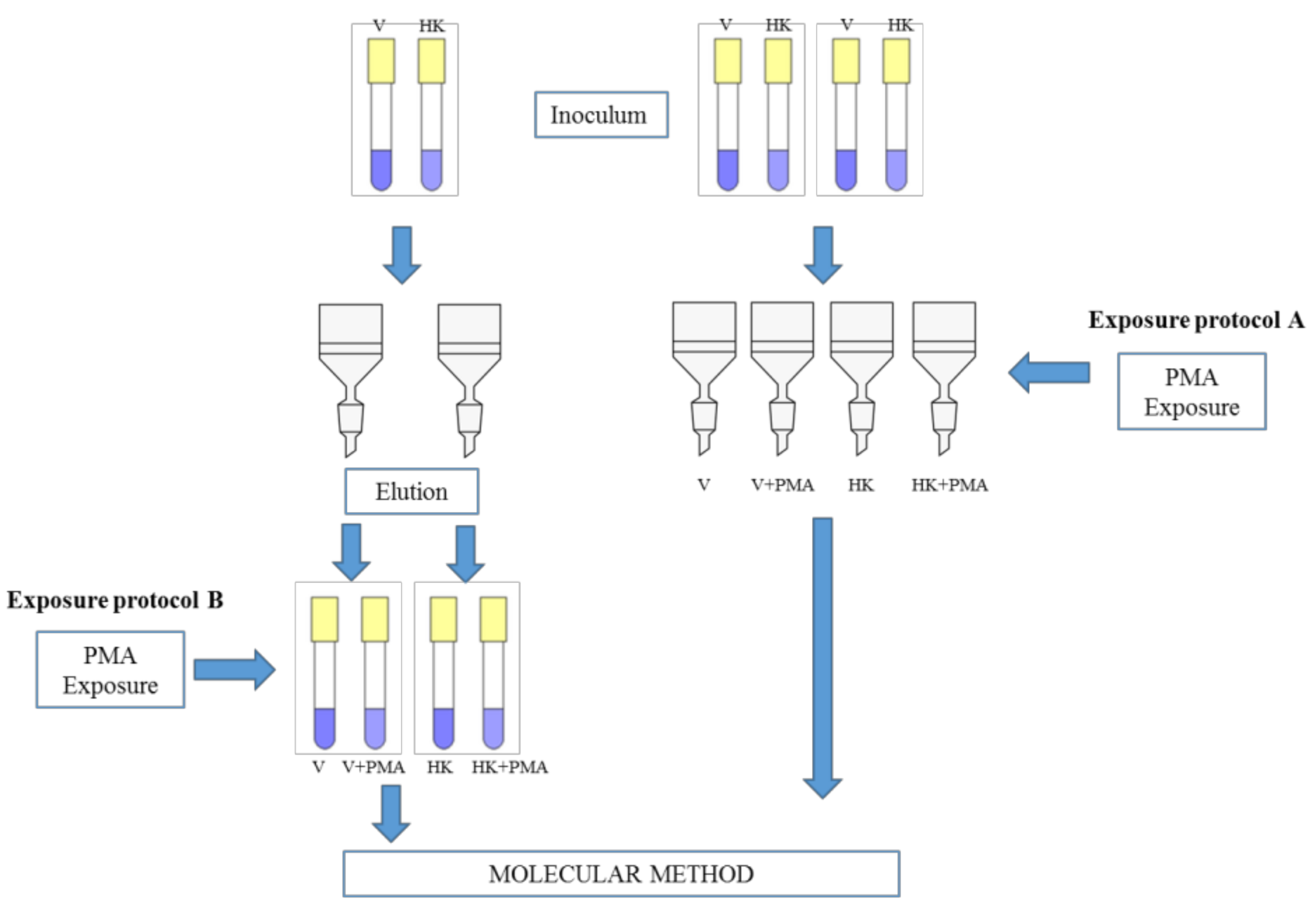
2.3. Exposure Protocols and Quantification by qPCR and PMA-qPCR
2.4. Statistical Analysis
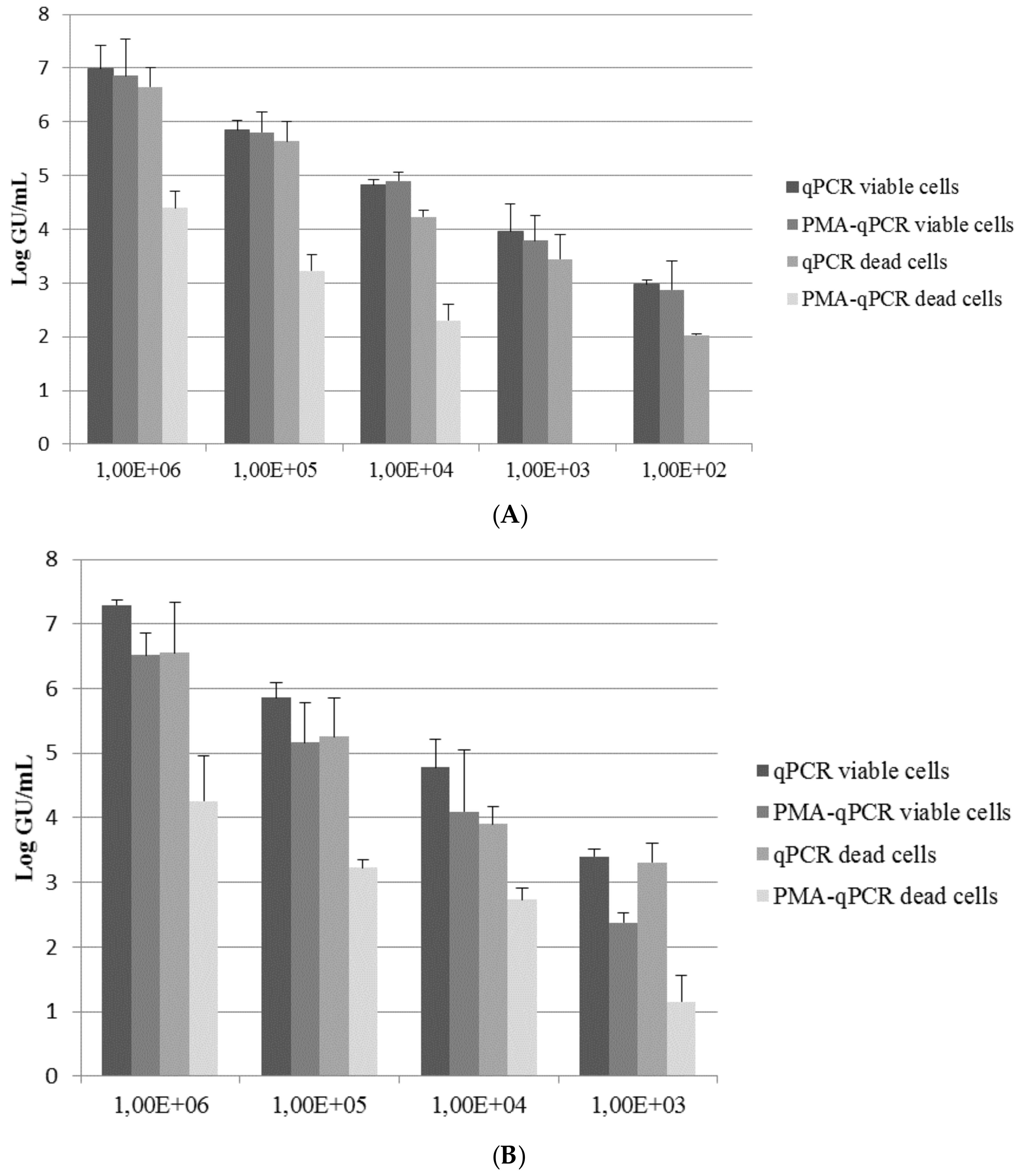
3. Results and Discussion
4. Conclusions
Author Contributions
Conflicts of Interest
References
- Bonetta, S.; Bonetta, S.; Ferretti, E.; Balocco, F.; Carraro, E. Evaluation of Legionella pneumophila contamination in Italian hotel water systems by quantitative real-time PCR and culture methods. J. Appl. Microbiol. 2010, 108, 1576–1583. [Google Scholar] [CrossRef] [PubMed]
- Ditommaso, S.; Giacomuzzi, M.; Ricciardi, E.; Zotti, C.M. Cultural and Molecular Evidence of Legionella spp. colonization in dental unit waterlines: Which is the best method for risk assessment? Int. J. Environ. Res. Public Health 2016, 13, 211. [Google Scholar] [CrossRef] [PubMed]
- Mansi, A.; Amori, I.; Marchesi, I.; Marcelloni, A.M.; Proietto, A.R.; Ferranti, G.; Magini, V.; Valeriani, F.; Borella, P. Legionella spp. survival after different disinfection procedures: Comparison between conventional culture, qPCR and EMA–qPCR. Microchem. J. 2014, 112, 65–69. [Google Scholar] [CrossRef]
- Walser, S.M.; Gerstner, D.G.; Brenner, B.; Höller, C.; Liebl, B.; Herr, C.E. Assessing the environmental health relevance of cooling towers—A systematic review of legionellosis outbreaks. Int. J. Hyg. Environ. Health 2014, 217, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Cunha, B.A.; Burillo, A.; Bouza, E. Legionnaires’ disease. Lancet 2016, 387, 376–385. [Google Scholar] [CrossRef]
- Yáñez, M.A.; Nocker, A.; Soria-Soria, E.; Múrtula, R.; Martínez, L.; Catalán, V. Quantification of viable Legionella pneumophila cells using propidium monoazide combined with quantitative PCR. J. Microbiol. Methods 2011, 85, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Ditommaso, S.; Ricciardi, E.; Giacomuzzi, M.; Arauco Rivera, S.R.; Ceccarelli, A.; Zotti, C.M. Overestimation of the Legionella spp. load in environmental samples by quantitative real-time PCR: Pretreatment with propidium monoazide as a tool for the assessment of an association between Legionella concentration and sanitary risk. Diagn. Microbiol. Infect. Dis. 2014, 80, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Fittipaldi, M.; Nocker, A.; Codony, F. Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods 2012, 91, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Viscogliosi, P.; Solignac, L.; Delattre, J. Viability PCR, a culture-independent method for rapid and selective quantification of viable Legionella pneumophila cells in environmental water samples. Appl. Environ. Microbiol. 2015, 75, 3502–3512. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Cheung, C.Y.; Camper, A.K. Comparison of propidium monoazide with ethidium monoazidefor differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Scaturro, M.; Fontana, S.; Dell’eva, I.; Helfer, P.; Marchio, M.; Stefanetti, M.V.; Cavallaro, M.; Miglietta, M.; Montagna, M.T.; De Giglio, O.; et al. A multicenter study of viable PCR using propidium monoazide to detect Legionella in water samples. Diagn. Microbiol. Infect. Dis. 2016, 85, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xin, H.; Li, S.F.Y. Multiplex PMA-qPCR assay with internal amplification control for simultaneous detection of viable Legionella pneumophila, Salmonella typhimurium, and Staphylococcus aureus in environmental waters. Environ. Sci. Technol. 2015, 49, 14249–14256. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.; Taguri, T.; Sugiyama, K.; Amemura-Maekawa, J.; Kura, F.; Watanabe, H. Comparison of ethidium monoazide and propidium monoazide for the selective detection of viable Legionella cells. Jpn. J. Infect. Dis. 2010, 63, 119–123. [Google Scholar] [PubMed]
- Ditommaso, S.; Giacomuzzi, M.; Ricciardi, E.; Zotti, C.M. Viability-qPCR for detecting Legionella: Comparison of two assays based on different amplicon lengths. Mol. Cell. Probes 2015, 29, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Ditommaso, S.; Giacomuzzi, M.; Ricciardi, E.; Arauco Rivera, S.R. Legionella in water samples: How can you interpret the results obtained by quantitative PCR? Mol. Cell. Probes 2015, 29, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Slimani, S.; Robyns, A.; Jarraud, S.; Molmeret, M.; Dusserre, E.; Mazure, C.; Facon, J.; Lina, G.; Etienne, J.; Ginevra, C. Evaluation of propidium monoazide (PMA) treatment directly on membrane filter for the enumeration of viable but non cultivable Legionella by qPCR. J. Microbiol. Methods 2012, 88, 319–321. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Breidt, F. Enumeration of viable Listeria monocytogenes cells by real-time PCR with propidium monoazide and ethidium monoazide in the presence of dead cells. Appl. Environ. Microbiol. 2007, 73, 8028–8031. [Google Scholar] [CrossRef] [PubMed]
- Kralik, P.; Nocker, A.; Pavlik, I. Mycobacterium avium subsp. Paratubercolosis viability determination using F57 quantitative PCR in combination with propidium monoazide treatment. Int. J. Food Microbiol. 2010, 141, S80–S86. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.T.; Chang, C.W. Rapid quantification of viable legionellae in water and biofilm using ethidium monoazide coupled with real-time quantitative PCR. J. Appl. Microbiol. 2010, 109, 623–634. [Google Scholar] [CrossRef] [PubMed]
| 7 µM | 12.5 µM | 25 µM | ||||
|---|---|---|---|---|---|---|
| −PMA | +PMA | −PMA | +PMA | −PMA | +PMA | |
| Protocol A (Filter) | 6.04 ± 0.15 | 5.89 ± 0.14 a | 5.97 ± 0.10 | 5.84 ± 0.18 a | 5.25 ± 0.11 | 5.14 ± 0.12 a |
| Legionella Reduction * | −0.15 | −0.13 | −0.11 | |||
| Protocol B (Liquid) | 6.41 ± 0.22 | 6.13 ± 0.20 a | 6.56 ± 0.18 | 6.23 ± 0.21 a | 6.66 ± 0.13 | 6.40 ± 0.38 a |
| Legionella Reduction * | −0.28 | −0.33 | −0.26 | |||
| Inoculum Concentration | PMA-Induced qPCR Signal Reduction a | |
|---|---|---|
| Exposure Method A (Filter) | Exposure Method B (Liquid) | |
| 1 × 106 | 2.25 | 2.29 |
| 1 × 105 | 2.42 | 2.04 |
| 1 × 104 | 1.93 * | 1.17 * |
| 1 × 103 | 3.45 * | 2.16 * |
| 1 × 102 | 2.03 | nv |
| Mean (SD) | 2.42 (0.61) | 1.91 (0.51) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonetta, S.; Pignata, C.; Bonetta, S.; Meucci, L.; Giacosa, D.; Marino, E.; Gilli, G.; Carraro, E. Viability of Legionella pneumophila in Water Samples: A Comparison of Propidium Monoazide (PMA) Treatment on Membrane Filters and in Liquid. Int. J. Environ. Res. Public Health 2017, 14, 467. https://doi.org/10.3390/ijerph14050467
Bonetta S, Pignata C, Bonetta S, Meucci L, Giacosa D, Marino E, Gilli G, Carraro E. Viability of Legionella pneumophila in Water Samples: A Comparison of Propidium Monoazide (PMA) Treatment on Membrane Filters and in Liquid. International Journal of Environmental Research and Public Health. 2017; 14(5):467. https://doi.org/10.3390/ijerph14050467
Chicago/Turabian StyleBonetta, Sara, Cristina Pignata, Silvia Bonetta, Lorenza Meucci, Donatella Giacosa, Elena Marino, Giorgio Gilli, and Elisabetta Carraro. 2017. "Viability of Legionella pneumophila in Water Samples: A Comparison of Propidium Monoazide (PMA) Treatment on Membrane Filters and in Liquid" International Journal of Environmental Research and Public Health 14, no. 5: 467. https://doi.org/10.3390/ijerph14050467






