A qPCR-Based Tool to Diagnose the Presence of Harmful Cyanobacteria and Cyanotoxins in Drinking Water Sources
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cyanobacterial Strains and Culturing
2.2. Study Sites
2.3. Cell Enumeration
2.4. Extraction of DNA from Cyanobacterial Cells
2.5. Duplex qPCR System
2.6. PCR Amplification, Cloning, and Standard Curves
2.7. Test of Inhibition on Gene Detection in Duplex Systems
2.8. Detection of Cyanotoxins
2.9. Statistical Analysis
3. Results and Discussion
3.1. Method Development for Quantification of Duplex qPCR Systems
3.1.1. Standard Curves
3.1.2. Influence of Gene Abundance in the Duplex Systems
3.2. Applications of the Duplex Systems in Reservoir Water Samples
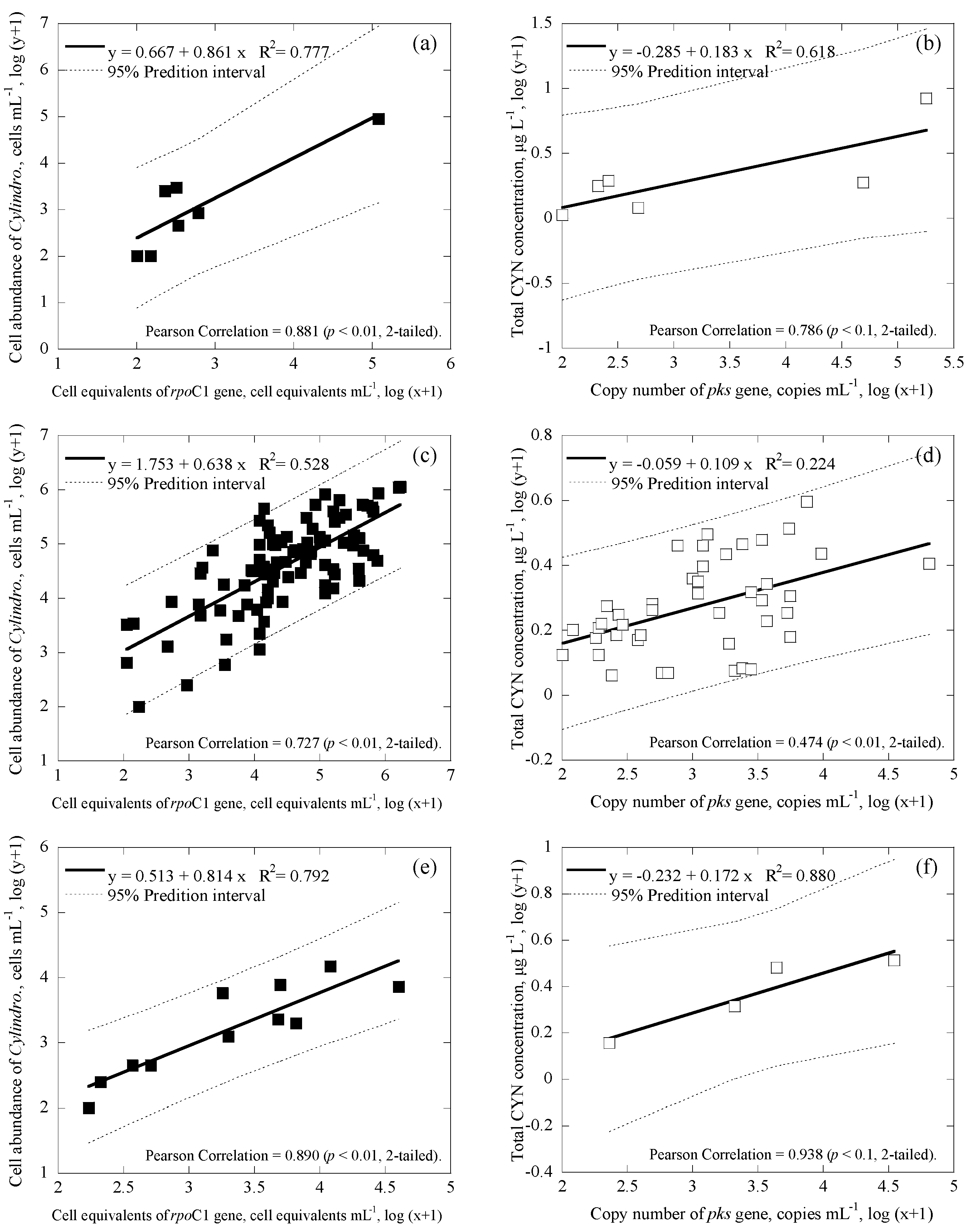
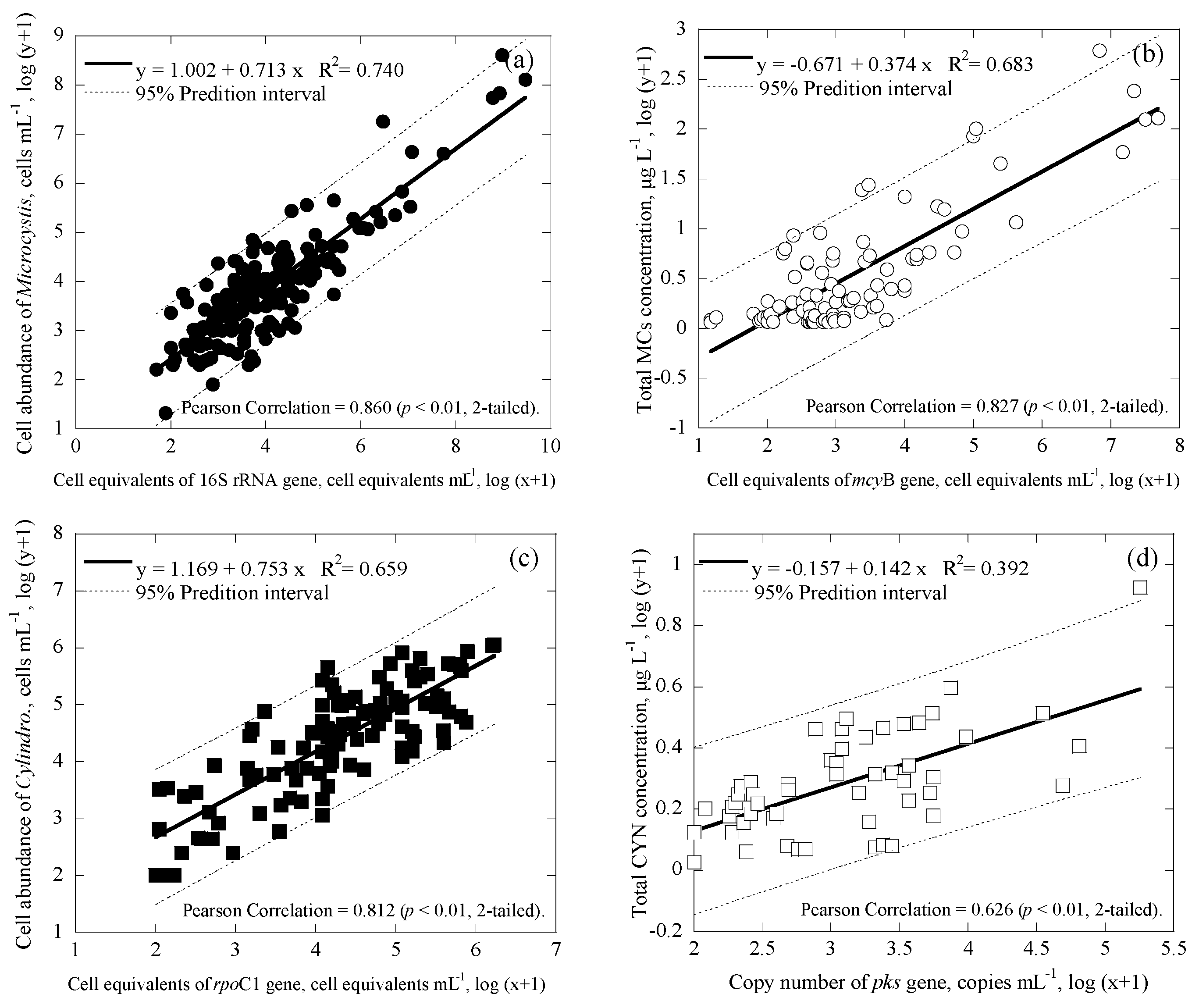
3.3. Correlations among Cell Numbers, Toxin Concentrations, Gene Copies and Cell Equivalents of the Samples
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Paerl, H.W.; Fulton, R.S.; Moisander, P.H.; Dyble, J. Harmful freshwater algal blooms, with an emphasis on cyanobacteria. Sci. World J. 2001, 1, 76–113. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, W.W. Cyanobacteria secondary metabolites—The cyanotoxins. J. Appl. Bacteriol. 1992, 72, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, W.W. Health effects of toxin-producing cyanobacteria:“The cyanohabs”. Hum. Ecol. Risk Assess. Int. J. 2001, 7, 1393–1407. [Google Scholar] [CrossRef]
- Yen, H.-K.; Lin, T.-F.; Tseng, I.-C. Detection and quantification of major toxigenic Microcystis genotypes in moo-tan reservoir and associated water treatment plant. J. Environ. Monit. 2012, 14, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Marbun, Y.R.; Yen, H.-K.; Lin, T.-F.; Lin, H.-L.; Michinaka, A. Rapid on-site monitoring of cylindrospermopsin-producers in reservoirs using quantitative PCR. Sustain. Environ. Res. 2012, 22, 143–151. [Google Scholar]
- Yen, H.; Lin, T.; Tseng, I.; Su, Y. Cyanobacteria toxins and toxin producers in nine drinking water reservoirs in taiwan. Water Sci. Technol. Water Supply 2006, 6, 161–167. [Google Scholar]
- Chapman, A.D.; Schelske, C.L. Recent appearance of Cylindrospemopsis (cyanobacteiua) in five hypereutrophic florida lakes1. J. Phycol. 1997, 33, 191–195. [Google Scholar] [CrossRef]
- Padisák, J. Cylindrospermopsis raciborskii (woloszynska) seenayya et subba raju, an expanding, highly adaptive cyanobacterium: Worldwide distribution and review of its ecology. Archiv Für Hydrobiologie Supplementband Monographische Beitrage 1997, 107, 563–593. [Google Scholar]
- Vaitomaa, J.; Rantala, A.; Halinen, K.; Rouhiainen, L.; Tallberg, P.; Mokelke, L.; Sivonen, K. Quantitative real-time PCR for determination of microcystin synthetase e copy numbers for Microcystis and Anabaena in lakes. Appl. Environ. Microbiol. 2003, 69, 7289–7297. [Google Scholar] [CrossRef] [PubMed]
- Valério, E.; Chambel, L.; Paulino, S.; Faria, N.; Pereira, P.; Tenreiro, R. Multiplex PCR for detection of microcystins-producing cyanobacteria from freshwater samples. Environ. Toxicol. 2010, 25, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lou, I.; Ung, W.K.; Kong, Y.; Mok, K.M. Application of PCR and real-time PCR for monitoring cyanobacteria, Microcystis spp. And Cylindrospermopsis raciborskii in macau freshwater reservoir. Front. Earth Sci. 2014, 8, 291–301. [Google Scholar] [CrossRef]
- Codd, G.A. Cyanobacterial toxins, the perception of water quality, and the prioritisation of eutrophication control. Ecol. Eng. 2000, 16, 51–60. [Google Scholar] [CrossRef]
- Duy, T.N.; Lam, P.K.; Shaw, G.R.; Connell, D.W. Toxicology and risk assessment of freshwater cyanobacterial (blue-green algal) toxins in water. In Reviews of Environmental Contamination and Toxicology; Springer: New York, NY, USA, 2000; pp. 113–185. [Google Scholar]
- Jochimsen, E.M.; Carmichael, W.W.; An, J.; Cardo, D.M.; Cookson, S.T.; Holmes, C.E.; Antunes, M.B.; de Melo Filho, D.A.; Lyra, T.M.; Barreto, V.S.T. Liver failure and death after exposure to microcystins at a hemodialysis center in brazil. N. Engl. J. Med. 1998, 338, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Runnegar, M.T.; Kong, S.M.; Zhong, Y.Z.; Ge, J.L.; Lu, S.C. The role of glutathione in the toxicity of a novel cyanobacterial alkaloid cylindrospermopsin in cultured rat hepatocytes. Biochem. Biophys. Res. Commun. 1994, 201, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Runnegar, M.T.; Kong, S.-M.; Zhong, Y.-Z.; Lu, S.C. Inhibition of reduced glutathione synthesis by cyanobacterial alkaloid cylindrospermopsin in cultured rat hepatocytes. Biochem. Pharmacol. 1995, 49, 219–225. [Google Scholar] [CrossRef]
- Kiss, T.; Vehovszky, Á.; Hiripi, L.; Kovács, A.; Vörös, L. Membrane effects of toxins isolated from a cyanobacterium, Cylindrospermopsis raciborskii, on identified molluscan neurones. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2002, 131, 167–176. [Google Scholar] [CrossRef]
- Saker, M.; Thomas, A.; Norton, J. Cattle mortality attributed to the toxic cyanobacterium Cylindrospermopsis raciborskii in an outback region of north queensland. Environ. Toxicol. 1999, 14, 179–182. [Google Scholar] [CrossRef]
- Sivonen, K.; Namikoshi, M.; Evans, W.R.; Carmichael, W.W.; Sun, F.; Rouhiainen, L.; Luukkainen, R.; Rinehart, K.L. Isolation and characterization of a variety of microcystins from 7 strains of the cyanobacterial genus Anabaena. Appl. Environ. Microbiol. 1992, 58, 2495–2500. [Google Scholar] [PubMed]
- Mohamed, Z.A.; Al Shehri, A.M. Microcystin-producing blooms of Anabaenopsis arnoldi in a potable mountain lake in saudi arabia. FEMS Microbiol. Ecol. 2009, 69, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Domingos, P.; Rubim, T.K.; Molica, R.J.R.; Azevedo, S.; Carmichael, W.W. First report of microcystin production by picoplanktonic cyanobacteria isolated from a northeast brazilian drinking water supply. Environ. Toxicol. 1999, 14, 31–35. [Google Scholar] [CrossRef]
- Dawson, R.M. The toxicology of microcystins. Toxicon 1998, 36, 953–962. [Google Scholar] [CrossRef]
- Sivonen, K.; Namikoshi, M.; Evans, W.R.; Fardig, M.; Carmichael, W.W.; Rinehart, K.L. Three new microcystins, cyclic heptapeptide hepatotoxins, from Nostoc sp. strain 152. Chem. Res. Toxicol. 1992, 5, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Luukkainen, R.; Sivonen, K.; Namikoshi, M.; Fardig, M.; Rinehart, K.L.; Niemela, S.I. Isolation and identification of 8 microcystins from 13 Oscillatoria-agardhii strains and structure of a new microcystin. Appl. Environ. Microbiol. 1993, 59, 2204–2209. [Google Scholar] [PubMed]
- Christiansen, G.; Fastner, J.; Erhard, M.; Borner, T.; Dittmann, E. Microcystin biosynthesis in Planktothrix: Genes, evolution, and manipulation. J. Bacteriol. 2003, 185, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Schembri, M.A.; Neilan, B.A.; Saint, C.P. Identification of genes implicated in toxin production in the cyanobacterium Cylindrospermopsis raciborskii. Environ. Toxicol. 2001, 16, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Spoof, L.; Berg, K.A.; Rapala, J.; Lahti, K.; Lepistö, L.; Metcalf, J.S.; Codd, G.A.; Meriluoto, J. First observation of cylindrospermopsin in Anabaena lapponica isolated from the boreal environment (Finland). Environ. Toxicol. 2006, 21, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Rücker, J.; Stüken, A.; Nixdorf, B.; Fastner, J.; Chorus, I.; Wiedner, C. Concentrations of particulate and dissolved cylindrospermopsin in 21 Aphanizomenon-dominated temperate lakes. Toxicon 2007, 50, 800–809. [Google Scholar] [CrossRef] [PubMed]
- Preußel, K.; Stüken, A.; Wiedner, C.; Chorus, I.; Fastner, J. First report on cylindrospermopsin producing Aphanizomenon flos-aquae (cyanobacteria) isolated from two german lakes. Toxicon 2006, 47, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.R.; Sukenik, A.; Livne, A.; Chiswell, R.K.; Smith, M.J.; Seawright, A.A.; Norris, R.L.; Eaglesham, G.K.; Moore, M.R. Blooms of the cylindrospermopsin containing cyanobacterium, Aphanizomenon ovalisporum (forti), in newly constructed lakes, queensland, australia. Environ. Toxicol. 1999, 14, 167–177. [Google Scholar] [CrossRef]
- Banker, R.; Carmeli, S.; Hadas, O.; Teltsch, B.; Porat, R.; Sukenik, A. Identification of cylindrospermopsin in Aphanizomenon ovalisporum (cyanophyceae) isolated from lake kinneret, israel1. J. Phycol. 1997, 33, 613–616. [Google Scholar] [CrossRef]
- Quesada, A.; Moreno, E.; Carrasco, D.; Paniagua, T.; Wormer, L.; Hoyos, C.d.; Sukenik, A. Toxicity of Aphanizomenon ovalisporum (cyanobacteria) in a Spanish water reservoir. Eur. J. Phycol. 2006, 41, 39–45. [Google Scholar] [CrossRef]
- Ohtani, I.; Moore, R.E.; Runnegar, M.T.C. Cylindrospermopsin: A potent hepatotoxin from the blue-green alga Cylindrospermopsis raciborskii. J. Am. Chem. Soc. 1992, 114, 7941–7942. [Google Scholar] [CrossRef]
- Li, R.; Carmichael, W.W.; Brittain, S.; Eaglesham, G.K.; Shaw, G.R.; Mahakhant, A.; Noparatnaraporn, N.; Yongmanitchai, W.; Kaya, K.; Watanabe, M.M. Isolation and identification of the cyanotoxin cylindrospermopsin and deoxy-cylindrospermopsin from a Thailand strain of Cylindrospermopsis raciborskii (cyanobacteria). Toxicon 2001, 39, 973–980. [Google Scholar] [CrossRef]
- Li, R.; Carmichael, W.W.; Brittain, S.; Eaglesham, G.K.; Shaw, G.R.; Liu, Y.; Watanabe, M.M. First report of the cyanotoxins cylindrospermopsin and deoxycylindrospermopsin from Raphidiopsis curvata (cyanobacteria). J. Phycol. 2001, 37, 1121–1126. [Google Scholar] [CrossRef]
- Harada, K.-i.; Ohtani, I.; Iwamoto, K.; Suzuki, M.; Watanabe, M.F.; Watanabe, M.; Terao, K. Isolation of cylindrospermopsin from a cyanobacterium Umezakia natans and its screening method. Toxicon 1994, 32, 73–84. [Google Scholar] [CrossRef]
- Neilan, B.; Saker, M.; Fastner, J.; Törökné, A.; Burns, B. Phylogeography of the invasive cyanobacterium Cylindrospermopsis raciborskii. Mol. Ecol. 2003, 12, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Organization, W.H. Guidelines for Drinking-Water Quality; World Health Organization: Geneva, Switzerland, 2004; Volume 1. [Google Scholar]
- Otsuka, S.; Suda, S.; Li, R.; Watanabe, M.; Oyaizu, H.; Matsumoto, S.; Watanabe, M.M. Phylogenetic relationships between toxic and non-toxic strains of the genus Microcystis based on 16S to 23S internal transcribed spacer sequence. FEMS Microbiol. Lett. 1999, 172, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Fastner, J.; Erhard, M.; von Döhren, H. Determination of oligopeptide diversity within a natural population of Microcystis spp. (cyanobacteria) by typing single colonies by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 2001, 67, 5069–5076. [Google Scholar] [CrossRef] [PubMed]
- Kurmayer, R.; Dittmann, E.; Fastner, J.; Chorus, I. Diversity of microcystin genes within a population of the toxic cyanobacterium Microcystis spp. In lake wannsee (Berlin, Germany). Microb. Ecol. 2002, 43, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.E.; Sarnelle, O.; Tillmanns, A.R. Effects of cyanobacterial toxicity and morphology on the population growth of freshwater zooplankton: Meta-analyses of laboratory experiments. Limnol. Oceanogr. 2006, 51, 1915–1924. [Google Scholar] [CrossRef]
- Alster, A.; Kaplan-Levy, R.N.; Sukenik, A.; Zohary, T. Morphology and phylogeny of a non-toxic invasive Cylindrospermopsis raciborskii from a mediterranean lake. Hydrobiologia 2010, 639, 115–128. [Google Scholar] [CrossRef]
- Baker, J.A.; Entsch, B.; Neilan, B.A.; McKay, D.B. Monitoring changing toxigenicity of a cyanobacterial bloom by molecular methods. Appl. Environ. Microbiol. 2002, 68, 6070–6076. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, J.P.; Giglio, S.; Monis, P.T.; Campbell, R.J.; Saint, C.P. Development and field testing of a real-time PCR assay for cylindrospermopsin-producing cyanobacteria. J. Appl. Microbiol. 2008, 104, 1503–1515. [Google Scholar] [CrossRef] [PubMed]
- Michinaka, A.; Yen, H.K.; Chiu, Y.T.; Tsao, H.W.; Lin, T.F. Rapid on-site multiplex assays for total and toxigenic Microcystis using real-time PCR with microwave cell disruption. Water Sci. Technol. 2012, 66, 1247–1252. [Google Scholar] [CrossRef] [PubMed]
- Al-Tebrineh, J.; Gehringer, M.M.; Akcaalan, R.; Neilan, B.A. A new quantitative PCR assay for the detection of hepatotoxigenic cyanobacteria. Toxicon 2011, 57, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Chu, F.; Huang, X.; Wei, R. Enzyme-linked immunosorbent assay for microcystins in blue-green algal blooms. J. Assoc. Off. Anal. Chem. 1989, 73, 451–456. [Google Scholar]
- Fischer, W.J.; Garthwaite, I.; Miles, C.O.; Ross, K.M.; Aggen, J.B.; Chamberlin, A.R.; Towers, N.R.; Dietrich, D.R. Congener-independent immunoassay for microcystins and nodularins. Environ. Sci. Technol. 2001, 35, 4849–4856. [Google Scholar] [CrossRef] [PubMed]
- Al-Tebrineh, J.; Pearson, L.A.; Yasar, S.A.; Neilan, B.A. A multiplex qPCR targeting hepato- and neurotoxigenic cyanobacteria of global significance. Harmful Algae 2012, 15, 19–25. [Google Scholar] [CrossRef]
- Te, S.H.; Chen, E.Y.; Gin, K.Y.-H. Comparison of quantitative PCR and droplet digital PCR multiplex assays for two genera of bloom-forming cyanobacteria, Cylindrospermopsis and Microcystis. Appl. Environ. Microbiol. 2015, 81, 5203–5211. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.; Sendall, B.C.; Gasser, R.B.; Menjivar, T.; Neilan, B.A.; Jex, A.R. Rapid, multiplex-tandem PCR assay for automated detection and differentiation of toxigenic cyanobacterial blooms. Mol. Cell. Probes 2013, 27, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Ngwa, F.F.; Madramootoo, C.A.; Jabaji, S. Development and application of a multiplex qPCR technique to detect multiple microcystin-producing cyanobacterial genera in a Canadian freshwater lake. J. Appl. Phycol. 2013, 26, 1675–1687. [Google Scholar] [CrossRef]
- Fortin, N.; Aranda-Rodriguez, R.; Jing, H.; Pick, F.; Bird, D.; Greer, C.W. Detection of microcystin-producing cyanobacteria in Missisquoi Bay, Quebec, Canada, using quantitative PCR. Appl. Environ. Microbiol. 2010, 76, 5105–5112. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, A.J.A.; Handy, S.M.; Wilhelm, S.W. Toxic Microcystis is widespread in lake erie: PCR detection of toxin genes and molecular characterization of associated cyanobacterial communities. Microb. Ecol. 2006, 51, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Rantala, A.; Rajaniemi-Wacklin, P.; Lyra, C.; Lepistö, L.; Rintala, J.; Mankiewicz-Boczek, J.; Sivonen, K. Detection of microcystin-producing cyanobacteria in finnish lakes with genus-specific microcystin synthetase gene E (mcyE) PCR and associations with environmental factors. Appl. Environ. Microbiol. 2006, 72, 6101–6110. [Google Scholar] [CrossRef] [PubMed]
- Koskenniemi, K.; Lyra, C.; Rajaniemi-Wacklin, P.; Jokela, J.; Sivonen, K. Quantitative real-time PCR detection of toxic Nodularia cyanobacteria in the baltic sea. Appl. Environ. Microbiol. 2007, 73, 2173–2179. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Y.; Mustapha, A. Rapid and simultaneous quantitation of Escherichia coli O157: H7, Salmonella, and Shigella in ground beef by multiplex real-time PCR and immunomagnetic separation. J. Food Prot. 2007, 70, 1366–1372. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, J.-Y.; Park, C.; Choi, I.-S.; Holt, P.S.; Seo, K.-H. Development of multiplex real-time PCR with internal amplification control for simultaneous detection of Salmonella and Cronobacter in powdered infant formula. Int. J. Food Microbiol. 2010, 144, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Peng, H.; Chen, W.; Cheng, J.; Wu, Y. Development of multiplex real-time PCR for simultaneous detection of three Potyviruses in tobacco plants. J. Appl. Microbiol. 2013, 114, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Theiss, W.C.; Carmichael, W.W.; Wyman, J.; Bruner, R. Blood pressure and hepatocellular effects of the cyclic heptapeptide toxin produced by the freshwater cyanobacterium (blue-green alga) Microcystis aeruginosa strain pcc-7820. Toxicon 1988, 26, 603–613. [Google Scholar] [CrossRef]
- Rippka, R. Isolation and purification of cyanobacteria. Meth. Enzymology 1988, 167, 3–27. [Google Scholar]
- Apha. Standard Methods for the Examination of Water and Wastewater; APHA: Washington, DC, USA, 1985. [Google Scholar]
- Chiu, Y.-T.; Yen, H.-K.; Lin, T.-F. An alternative method to quantify 2-MIB producing cyanobacteria in drinking water reservoirs: Method development and field applications. Environ. Res. 2016, 151, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Rinta-Kanto, J.M.; Ouellette, A.J.A.; Boyer, G.L.; Twiss, M.R.; Bridgeman, T.B.; Wilhelm, S.W. Quantification of toxic Microcystis spp. During the 2003 and 2004 blooms in western lake erie using quantitative real-time PCR. Environ. Sci. Technol. 2005, 39, 4198–4205. [Google Scholar] [CrossRef] [PubMed]
- Fergusson, K.M.; Saint, C.P. Multiplex PCR assay for Cylindrospermopsis raciborskii and cylindrospermopsin-producing cyanobacteria. Environ. Toxicol. 2003, 18, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.M.; Schembri, M.A.; Baker, P.D.; Saint, C.P. Molecular characterization of the toxic cyanobacterium Cylindrospermopsis raciborskii and design of a species-specific PCR. Appl. Environ. Microbiol. 2000, 66, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Kurmayer, R.; Kutzenberger, T. Application of real-time PCR for quantification of microcystin genotypes in a population of the toxic cyanobacterium Microcystis sp. Appl. Environ. Microbiol. 2003, 69, 6723–6730. [Google Scholar] [CrossRef] [PubMed]
- Griese, M.; Lange, C; Soppa, J. Ploidy in cyanobacteria. FEMS Microbiol. Lett. 2011, 323, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Chow, G.C. Tests of equality between sets of coefficients in two linear regressions. Econometrica 1960, 28, 591–605. [Google Scholar] [CrossRef]
- Elnifro, E.M.; Ashshi, A.M.; Cooper, R.J.; Klapper, P.E. Multiplex PCR: Optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000, 13, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Henegariu, O.; Heerema, N.A.; Dlouhy, S.R.; Vance, G.H.; Vogt, P.H. Multiplex PCR: Critical parameters and step-by-step protocol. Biotechniques 1997, 23, 504–511. [Google Scholar] [PubMed]
- Sint, D.; Raso, L.; Traugott, M. Advances in multiplex PCR: Balancing primer efficiencies and improving detection success. Methods Ecol. Evolut. 2012, 3, 898–905. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.G.; Connell, C.R.; Bloch, W. Allelic discrimination by nick-translation PCR with fluorgenic probes. Nucleic Acids Res. 1993, 21, 3761–3766. [Google Scholar] [CrossRef] [PubMed]
- Oliver, R.L. Floating and sinking in gas-vacuolate cyanobacteria. J. Phycol. 1994, 30, 161–173. [Google Scholar] [CrossRef]
- Codd, G.A.; Morrison, L.F.; Metcalf, J.S. Cyanobacterial toxins: Risk management for health protection. Toxicol. Appl. Pharmacol. 2005, 203, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Kaebernick, M.; Rohrlack, T.; Christoffersen, K.; Neilan, B.A. A spontaneous mutant of microcystin biosynthesis: Genetic characterization and effect on Daphnia. Environ. Microbiol. 2001, 3, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Tillett, D.; Parker, D.L.; Neilan, B.A. Detection of toxigenicity by a probe for the microcystin synthetase a gene (mcyA) of the cyanobacterial genus Microcystis: Comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies. Appl. Environ. Microbiol. 2001, 67, 2810–2818. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, T.; Asayama, M.; Fujii, K.; Harada, K.; Shirai, M. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J. Biochem. 1999, 126, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Mikalsen, B.; Boison, G.; Skulberg, O.M.; Fastner, J.; Davies, W.; Gabrielsen, T.M.; Rudi, K.; Jakobsen, K.S. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 2003, 185, 2774–2785. [Google Scholar] [CrossRef] [PubMed]
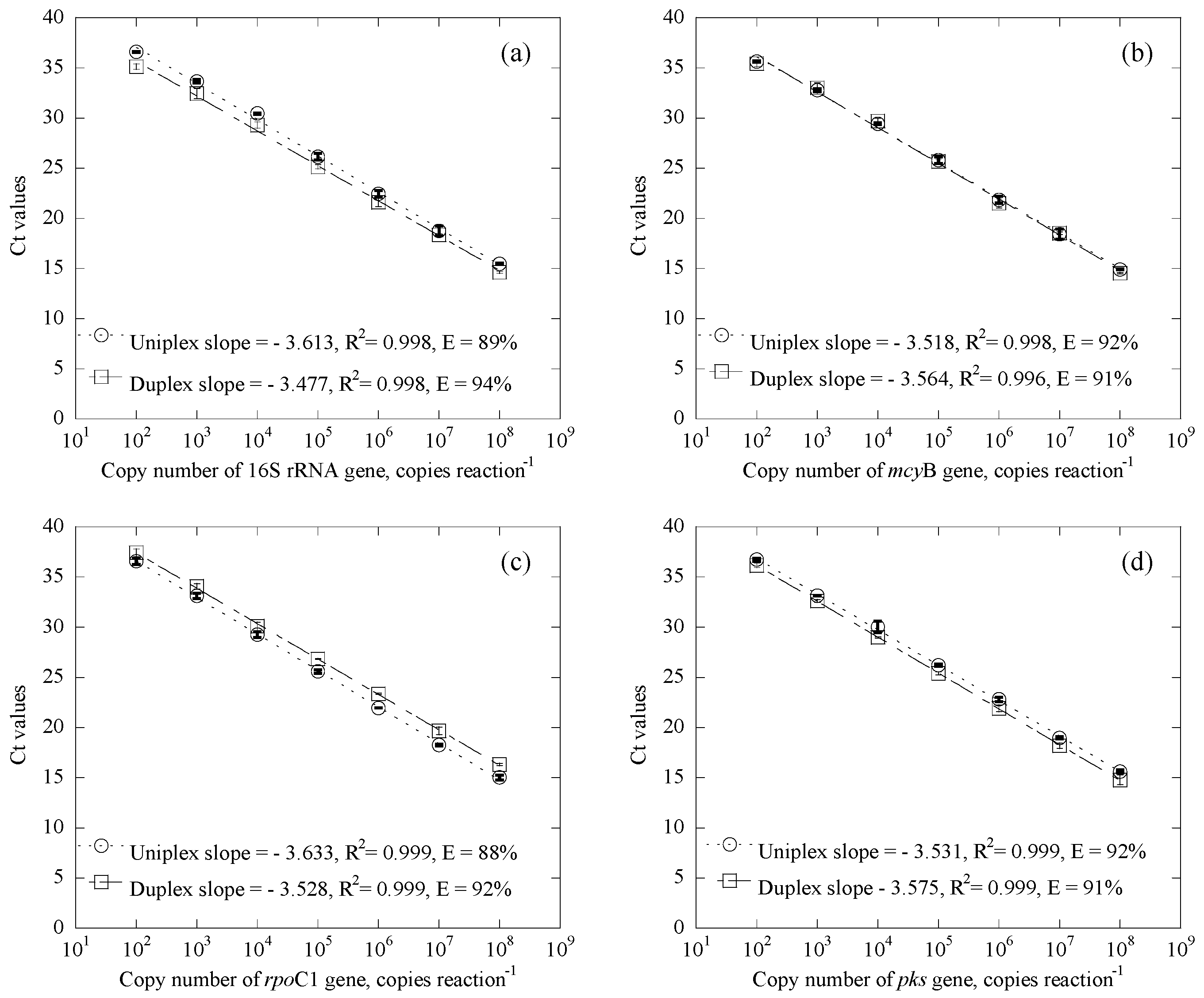
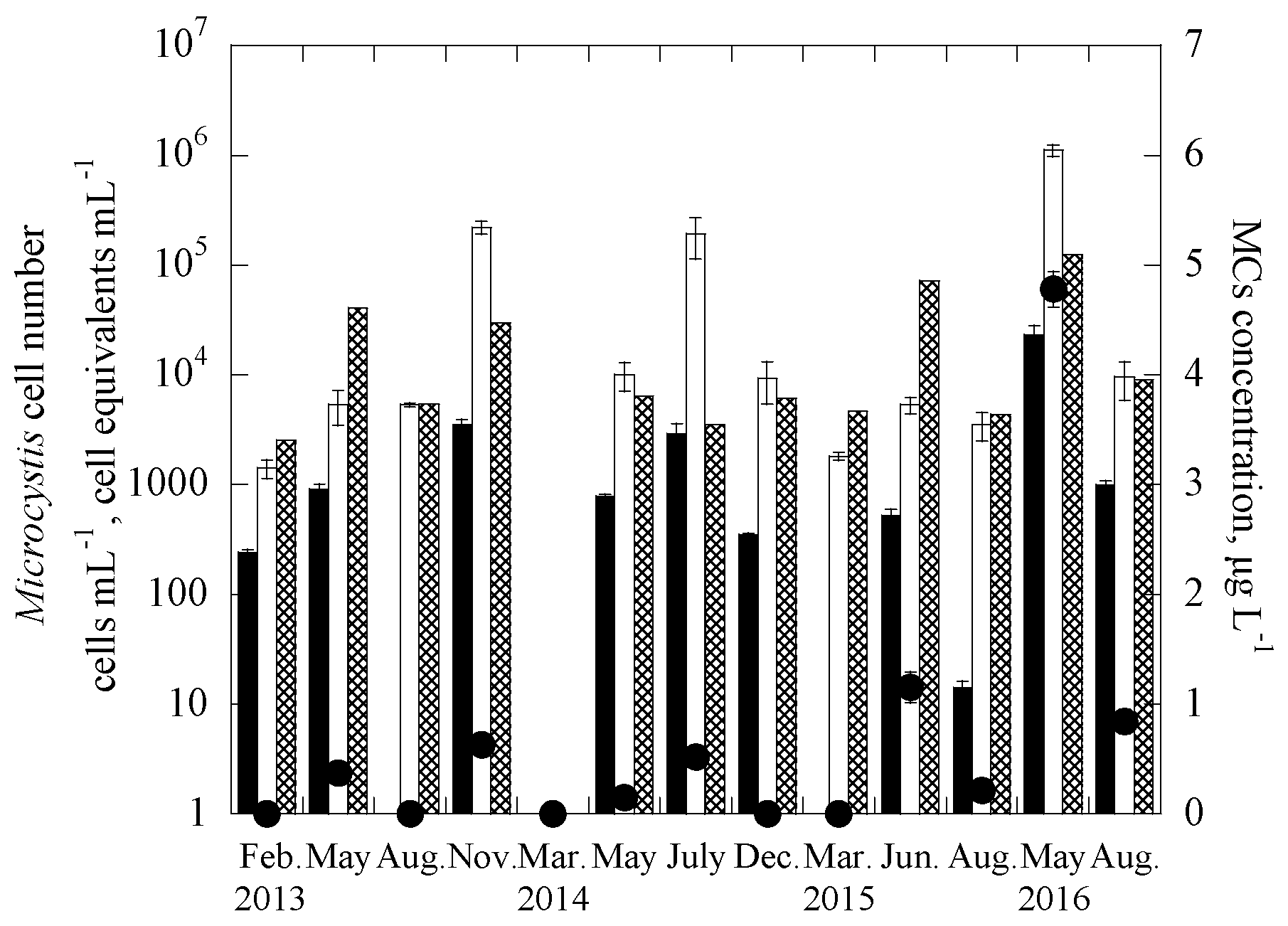
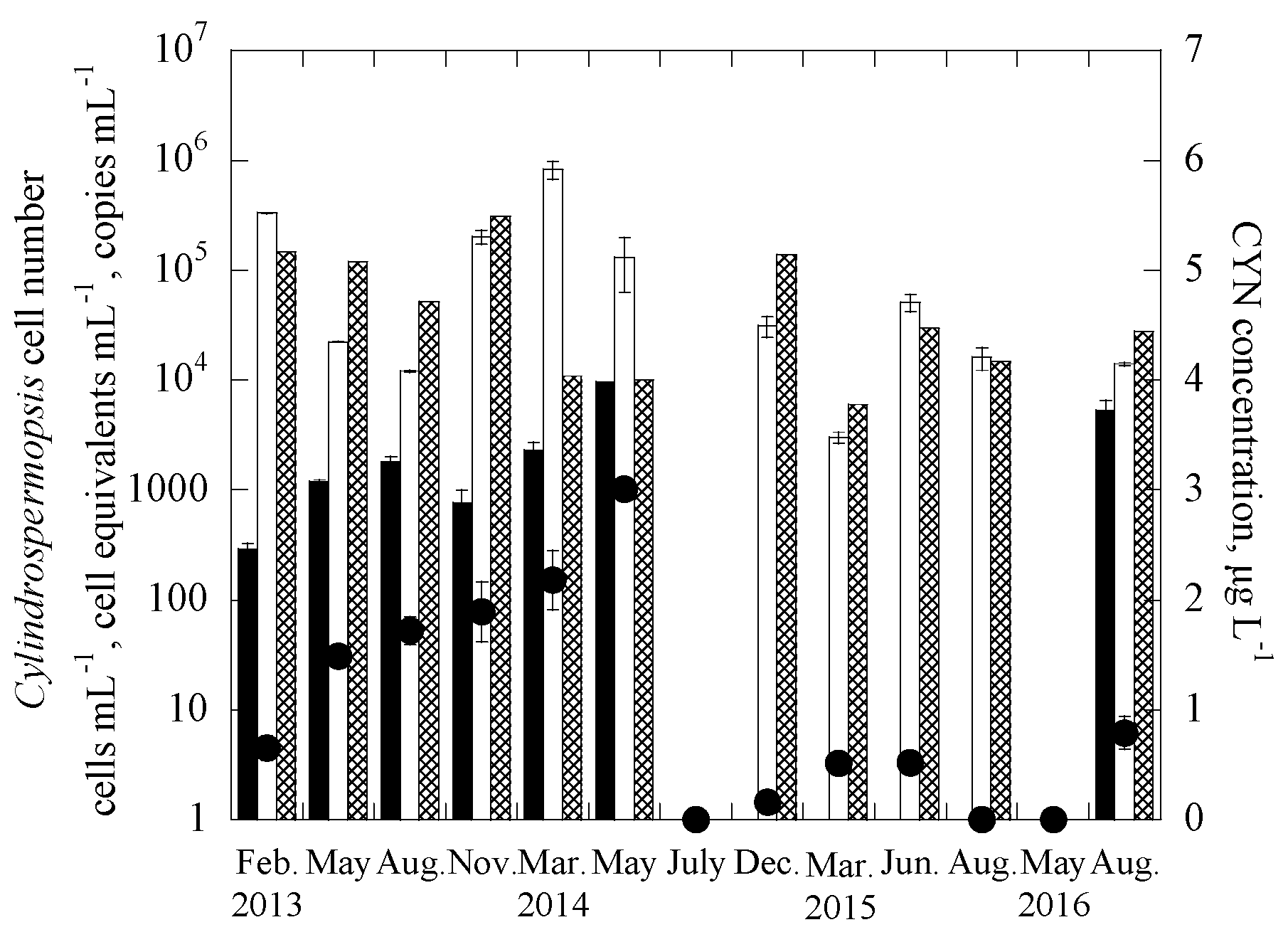
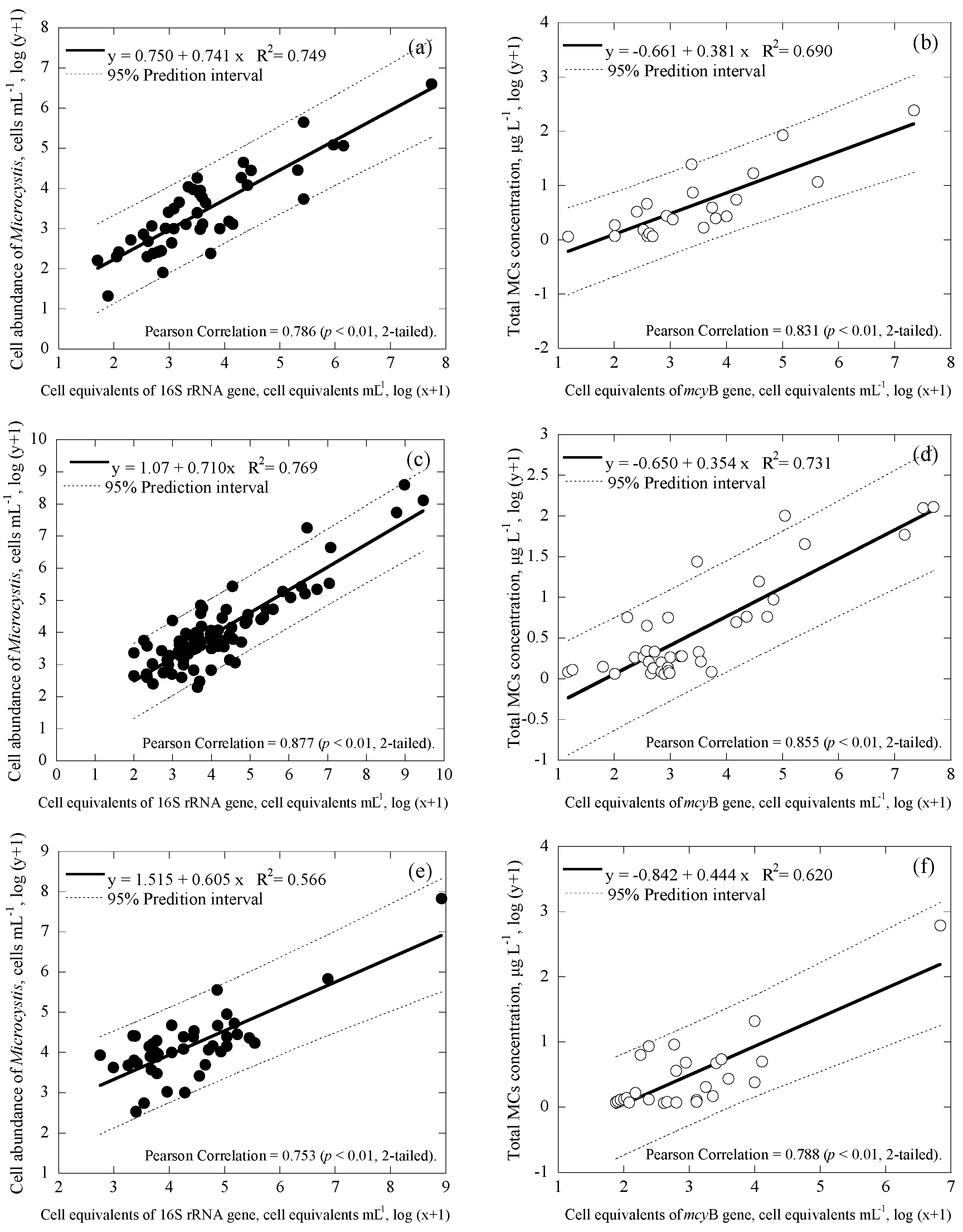
| Samples | Date | Uniplex (Ct Value) | Duplex (Ct Value) | CYN Concentration (μg/L) | |||
|---|---|---|---|---|---|---|---|
| pks Gene | rpoC1 Gene | pks Gene | rpoC1 Gene | ||||
| THR | 2013 | February | 38.64 (±0.14) 1 | 27.41 (±0.10) | - 2 | 28.48 (±0.06) | 0.65 |
| November | 37.13 (±0.17) | 28.27 (±0.17) | - | 29.26 (±0.05) | 1.89 | ||
| 2014 | March | 35.68 (±0.15) | 25.91 (±0.05) | - | 27.10 (±0.04) | 2.18 | |
| Duplex Microcystis System | Duplex Cylindrospermopsis System | ||||||
|---|---|---|---|---|---|---|---|
| Copies/Reaction | Ct Value | Copies/Reaction | Ct Value | ||||
| mcyB | 16S rRNA | mcyB | 16S rRNA | pks | rpoC1 | pks | rpoC1 |
| 103 | 103 | 33.52 (±0.42)1 | 32.04 (±0.07) | 103 | 103 | 32.69 (±0.33) | 34.12 (±0.23) |
| 104 | 33.41 (±0.57) | 30.07 (±0.04) | 104 | 32.70 (±0.28) | 30.31 (±0.13) | ||
| 105 | 32.84 (±0.61) | 29.95 (±0.37) | 105 | - 2 | 26.69 (±0.32) | ||
| 106 | 33.40 (±0.08) | 21.68 (±0.42) | 106 | - | 23.39 (±0.08) | ||
| 107 | 32.69 (±0.43) | 18.29 (±0.26) | 107 | - | 19.99 (±0.04) | ||
| 104 | 103 | 29.91 (±0.01) | 32.33 (±0.36) | 104 | 103 | 29.07 (±0.45) | 33.51 (±0.06) |
| 104 | 29.75 (±0.41) | 29.66 (±0.17) | 104 | 29.05 (±0.16) | 30.82 (±0.11) | ||
| 105 | 30.50 (±0.38) | 26.03 (±0.06) | 105 | 28.86 (±0.13) | 26.62 (±0.62) | ||
| 106 | 30.31 (±0.30) | 22.19 (±0.01) | 106 | - | 23.28 (±0.24) | ||
| 107 | 30.01 (±0.29) | 18.55 (±0.57) | 107 | - | 19.69 (±0.18) | ||
| 105 | 103 | 26.13 (±0.21) | - | 105 | 103 | 25.75 (±0.07) | 33.29 (±0.15) |
| 104 | 26.23 (±0.26) | 29.81 (±0.48) | 104 | 25.73 (±0.13) | 30.78 (±0.28) | ||
| 105 | 26.12 (±0.40) | 25.69 (±0.52) | 105 | 25.43 (±0.08) | 27.32 (±0.07) | ||
| 106 | 25.85 (±0.33) | 21.92 (±0.10) | 106 | 24.96 (±0.25) | 24.16 (±0.13) | ||
| 107 | 25.85 (±0.31) | 18.53 (±0.47) | 107 | - | 19.76 (±0.13) | ||
| 106 | 103 | 22.37 (±0.04) | - | 106 | 103 | 22.05 (±0.28) | - |
| 104 | 22.31 (±0.48) | - | 104 | 21.99 (±0.07) | 30.59 (±0.20) | ||
| 105 | 22.54 (±0.54) | 25.54 (±0.04) | 105 | 22.20 (±0.05) | 27.41 (±0.55) | ||
| 106 | 21.88 (±0.62) | 21.31 (±0.00) | 106 | 22.39 (±0.13) | 23.96 (±0.25) | ||
| 107 | 22.00 (±0.60) | 18.18 (±0.73) | 107 | 21.81 (±0.18) | 19.87 (±0.13) | ||
| 107 | 103 | 18.88 (±0.13) | - | 107 | 103 | 18.35 (±0.01) | - |
| 104 | 19.03 (±0.06) | - | 104 | 18.44 (±0.11) | - | ||
| 105 | 18.94 (±0.50) | - | 105 | 18.37 (±0.07) | 26.91 (±0.31) | ||
| 106 | 18.73 (±0.22) | 21.95 (±0.33) | 106 | 18.45 (±0.30) | 24.13 (±0.03) | ||
| 107 | 18.78 (±0.20) | 18.38 (±0.11) | 107 | 18.24 (±0.13) | 19.88 (±0.14) | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiu, Y.-T.; Chen, Y.-H.; Wang, T.-S.; Yen, H.-K.; Lin, T.-F. A qPCR-Based Tool to Diagnose the Presence of Harmful Cyanobacteria and Cyanotoxins in Drinking Water Sources. Int. J. Environ. Res. Public Health 2017, 14, 547. https://doi.org/10.3390/ijerph14050547
Chiu Y-T, Chen Y-H, Wang T-S, Yen H-K, Lin T-F. A qPCR-Based Tool to Diagnose the Presence of Harmful Cyanobacteria and Cyanotoxins in Drinking Water Sources. International Journal of Environmental Research and Public Health. 2017; 14(5):547. https://doi.org/10.3390/ijerph14050547
Chicago/Turabian StyleChiu, Yi-Ting, Yi-Hsuan Chen, Ting-Shaun Wang, Hung-Kai Yen, and Tsair-Fuh Lin. 2017. "A qPCR-Based Tool to Diagnose the Presence of Harmful Cyanobacteria and Cyanotoxins in Drinking Water Sources" International Journal of Environmental Research and Public Health 14, no. 5: 547. https://doi.org/10.3390/ijerph14050547






