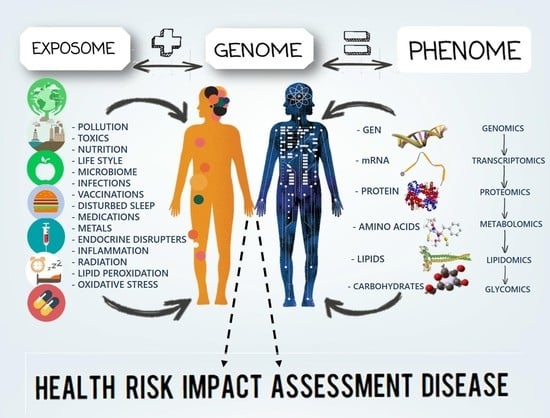
Mini-Review: The Contribution of Intermediate Phenotypes to GxE Effects on Disorders of Body Composition in the New OMICS Era
Abstract
:1. Introduction
2. Gene-Environment Interaction
3. Determinants of Unhealthy Eating Habits and Physical Inactivity: Risk Behaviors and Disorders of Body Composition as Health Consequences
4. Intermediate Phenotypes
5. Models and Mechanisms of Gene, Environment, and Behavior Interactions in Disease
6. Beyond Health Consequences: Deep Phenotyping for Early Biological Prevention
7. Beyond Risk Behaviors: The Exposome
8. Beyond Intermediate Phenotypes: Systems Genomics Methods
9. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rosen, C.J.; Klibanski, A. Bone, fat, and body composition: Evolving concepts in the pathogenesis of osteoporosis. Am. J. Med. 2009, 122, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Kawai, M.; de Paula, F.J.; Rosen, C.J. New insights into osteoporosis: The bone-fat connection. J. Int. Med. 2012, 272, 317–329. [Google Scholar] [CrossRef] [PubMed]
- de Paula, F.J.; Horowitz, M.C.; Rosen, C.J. Novel insights into the relationship between diabetes and osteoporosis. Diabetes Metab. Res. Rev. 2010, 26, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Spiegelman, B.M. What we talk about when we talk about fat. Cell 2014, 156, 20–44. [Google Scholar] [CrossRef] [PubMed]
- Cawthon, P.M. Gender differences in osteoporosis and fractures. Clin. Orthop. Relat. Res. 2011, 469, 1900–1905. [Google Scholar] [CrossRef] [PubMed]
- Jiao, H.; Xiao, E.; Graves, D.T. Diabetes and Its Effect on Bone and Fracture Healing. Curr. Osteoporos. Rep. 2015, 13, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Kremer, R.; Gilsanz, V. Fat and Bone: An Odd Couple. Front. Endocrinol. 2015, 6, 190. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.J. Gene-environment interactions in human diseases. Nat. Rev. Genet. 2005, 6, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Halldorsdottir, T.; Binder, E.B. Gene x Environment Interactions: From Molecular Mechanisms to Behavior. Annu. Rev. Psychol. 2017, 68, 215–241. [Google Scholar] [CrossRef] [PubMed]
- Manuck, S.B.; McCaffery, J.M. Gene-environment interaction. Annu. Rev. Psychol. 2014, 65, 41–70. [Google Scholar] [CrossRef] [PubMed]
- Te Pas, M.F.; Madsen, O.; Calus, M.P.; Smits, M.A. The Importance of Endophenotypes to Evaluate the Relationship between Genotype and External Phenotype. Int. J. Mol. Sci. 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Khoury, M.J.; Gwinn, M.; Clyne, M.; Yu, W. Genetic epidemiology with a capital E, ten years after. Genet. Epidemiol. 2011, 35, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.G.; Olden, K. Gene-environment interactions in the development of complex disease phenotypes. Int. J. Environ. Res. Public Health 2008, 5, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Pellmar, T.C.; Brandt, E.N., Jr.; Baird, M.A. Health and behavior: The interplay of biological, behavioral, and social influences: Summary of an Institute of Medicine report. Am. J. Health Promot. 2002, 16, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Rosen, C.J.; Bouxsein, M.L. Mechanisms of disease: Is osteoporosis the obesity of bone? Nat. Clin. Pract. Rheumatol. 2006, 2, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Townshend, T.; Lake, A. Obesogenic environments: Current evidence of the built and food environments. Perspect. Public Health 2017, 137, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, N.S.; Catenacci, V.A.; Wyatt, H.R.; Hill, J.O. Obesity: Overview of an epidemic. Psychiatr. Clin. North Am. 2011, 34, 717–732. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.H.; Venners, S.A.; Terwedow, H.A.; Feng, Y.; Niu, T.; Li, Z.; Laird, N.; Brain, J.D.; Cummings, S.R.; Bouxsein, M.L.; et al. Relation of body composition, fat mass, and serum lipids to osteoporotic fractures and bone mineral density in Chinese men and women. Am. J. Clin. Nutr. 2006, 83, 146–154. [Google Scholar] [PubMed]
- Zerwekh, J.E.; Ruml, L.A.; Gottschalk, F.; Pak, C.Y. The effects of twelve weeks of bed rest on bone histology, biochemical markers of bone turnover, and calcium homeostasis in eleven normal subjects. J. Bone Miner. Res. 1998, 13, 1594–1601. [Google Scholar] [CrossRef] [PubMed]
- Zillikens, M.C.; Uitterlinden, A.G.; van Leeuwen, J.P.; Berends, A.L.; Henneman, P.; van Dijk, K.W.; Oostra, B.A.; van Duijn, C.M.; Pols, H.A.; Rivadeneira, F.; et al. The role of body mass index, insulin, and adiponectin in the relation between fat distribution and bone mineral density. Calcif. Tissue Int. 2010, 86, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lim, S.Y.; Kim, J.H. Nutrient intake risk factors of osteoporosis in postmenopausal women. Asia Pac. J. Clin. Nutr. 2008, 17, 270–275. [Google Scholar] [PubMed]
- Fernandes, T.A.P.; Goncalves, L.M.L.; Brito, J.A.A. Relationships between Bone Turnover and Energy Metabolism. J. Diabetes Res. 2017, 2017, 9021314. [Google Scholar] [CrossRef] [PubMed]
- Shulman, J.M.; Chibnik, L.B.; Aubin, C.; Schneider, J.A.; Bennett, D.A.; De Jager, P.L. Intermediate phenotypes identify divergent pathways to Alzheimer’s disease. PLoS ONE 2010, 5, e11244. [Google Scholar] [CrossRef] [PubMed]
- Lenzenweger, M.F. Endophenotype, intermediate phenotype, biomarker: Definitions, concept comparisons, clarifications. Depress Anxiety 2013, 30, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Yamasue, H. Using endophenotypes to examine molecules related to candidate genes as novel therapeutics: The “endophenotype-associated surrogate endpoint (EASE)” concept. Neurosci. Res. 2015, 99, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lassere, M.N. The Biomarker-Surrogacy Evaluation Schema: a review of the biomarker-surrogate literature and a proposal for a criterion-based, quantitative, multidimensional hierarchical levels of evidence schema for evaluating the status of biomarkers as surrogate endpoints. Stat. Methods Med. Res. 2008, 17, 303–340. [Google Scholar] [PubMed]
- Deans, A.R.; Lewis, S.E.; Huala, E.; Anzaldo, S.S.; Ashburner, M.; Balhoff, J.P.; Blackburn, D.C.; Blake, J.A.; Burleigh, J.G.; Chanet, B.; et al. Finding our way through phenotypes. PLoS Biol. 2015, 13, e1002033. [Google Scholar] [CrossRef] [PubMed]
- Grigorenko, E.L. The inherent complexities of gene-environment interactions. J. Gerontol. B. Psychol. Sci. Soc. Sci. 2005, 60, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Rebbeck, T.R. The contribution of inherited genotype to breast cancer. Breast. Cancer Res. 2002, 4, 85–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastarrachea, R.A.; Gallegos-Cabriales, E.C.; Nava-Gonzalez, E.J.; Haack, K.; Voruganti, V.S.; Charlesworth, J.; Laviada-Molina, H.A.; Veloz-Garza, R.A.; Cardenas-Villarreal, V.M.; Valdovinos-Chavez, S.B.; et al. Integrating genomic analysis with the genetic basis of gene expression: Preliminary evidence of the identification of causal genes for cardiovascular and metabolic traits related to nutrition in Mexicans. Adv. Nutr. 2012, 3, 596S–604S. [Google Scholar] [CrossRef] [PubMed]
- Nava-Gonzalez, E.J.; Gallegos-Cabriales, E.C.; Bastarrachea, R.A. Phenotypes of bone and adipose tissue metabolism. A systematic review of their relationship. Rev. Med. Inst. Mex. Seguro Soc. 2014, 52, 644–650. [Google Scholar] [PubMed]
- Nava-Gonzalez, E.J.; Cerda-Flores, R.M.; Garcia-Hernandez, P.A.; Jasso-de la Pena, G.A.; Bastarrachea, R.A.; Gallegos-Cabriales, E.C. Bone mineral density and its association with body composition and metabolic biomarkers of insulin-glucose axis, bone and adipose tissue in women. Gac. Med. Mex. 2015, 151, 731–740. [Google Scholar] [PubMed]
- Meng, Q.; Makinen, V.P.; Luk, H.; Yang, X. Systems Biology Approaches and Applications in Obesity, Diabetes, and Cardiovascular Diseases. Curr. Cardiovasc. Risk Rep. 2013, 7, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, M.C.; Hu, F.B. Systems Epidemiology: A New Direction in Nutrition and Metabolic Disease Research. Curr. Nutr. Rep. 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Girirajan, S. Missing heritability and where to find it. Genome Biol. 2017, 18, 89. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.J.; Bosy-Westphal, A.; Krawczak, M. Genetic studies of common types of obesity: A critique of the current use of phenotypes. Obes. Rev. 2010, 11, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Haring, R.; Wallaschofski, H. Diving through the “-Omics”: The case for deep phenotyping and systems epidemiology. OMICS 2012, 16, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Tracy, R.P. ‘Deep phenotyping’: Characterizing populations in the era of genomics and systems biology. Curr. Opin. Lipidol. 2008, 19, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Secor, S.M. Specific dynamic action: A review of the postprandial metabolic response. J. Comp. Physiol. B 2009, 179, 1–56. [Google Scholar] [CrossRef] [PubMed]
- Delude, C.M. Deep phenotyping: The details of disease. Nature 2015, 527, S14–S15. [Google Scholar] [CrossRef] [PubMed]
- Stingone, J.A.; Buck Louis, G.M.; Nakayama, S.F.; Vermeulen, R.C.; Kwok, R.K.; Cui, Y.; Balshaw, D.M.; Teitelbaum, S.L. Toward Greater Implementation of the Exposome Research Paradigm within Environmental Epidemiology. Annu. Rev. Public Health 2017, 38, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Siroux, V.; Agier, L.; Slama, R. The exposome concept: A challenge and a potential driver for environmental health research. Eur. Respir. Rev. 2016, 25, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Vineis, P. A self-fulfilling prophecy: Are we underestimating the role of the environment in gene-environment interaction research? Int. J. Epidemiol. 2004, 33, 945–946. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, E.R.; Ritchie, M.D. Integrating heterogeneous high-throughput data for meta-dimensional pharmacogenomics and disease-related studies. Pharmacogenomics 2012, 13, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Suravajhala, P.; Kogelman, L.J.; Kadarmideen, H.N. Multi-omic data integration and analysis using systems genomics approaches: Methods and applications in animal production, health and welfare. Genet. Sel. Evol. 2016, 48, 38. [Google Scholar] [CrossRef] [PubMed]
- Gonnelli, S.; Caffarelli, C.; Nuti, R. Obesity and fracture risk. Clin. Cases Miner. Bone MeTable 2014, 11, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Jackuliak, P.; Payer, J. Osteoporosis, fractures, and diabetes. Int. J. Endocrinol. 2014, 2014, 820615. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Tandon, V.R.; Mahajan, S.; Mahajan, V.; Mahajan, A. Obesity: Friend or foe for osteoporosis. J. Midlife Health 2014, 5, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Candido, F.G.; Bressan, J. Vitamin, D. Link between osteoporosis, obesity, and diabetes? Int. J. Mol. Sci. 2014, 15, 6569–6591. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.; Lane, H.Y. Machine learning and systems genomics approaches for multi-omics data. Biomark. Res. 2017, 5, 2. [Google Scholar] [CrossRef] [PubMed]





© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nava-Gonzalez, E.J.; Gallegos-Cabriales, E.C.; Leal-Berumen, I.; Bastarrachea, R.A. Mini-Review: The Contribution of Intermediate Phenotypes to GxE Effects on Disorders of Body Composition in the New OMICS Era. Int. J. Environ. Res. Public Health 2017, 14, 1079. https://doi.org/10.3390/ijerph14091079
Nava-Gonzalez EJ, Gallegos-Cabriales EC, Leal-Berumen I, Bastarrachea RA. Mini-Review: The Contribution of Intermediate Phenotypes to GxE Effects on Disorders of Body Composition in the New OMICS Era. International Journal of Environmental Research and Public Health. 2017; 14(9):1079. https://doi.org/10.3390/ijerph14091079
Chicago/Turabian StyleNava-Gonzalez, Edna J., Esther C. Gallegos-Cabriales, Irene Leal-Berumen, and Raul A. Bastarrachea. 2017. "Mini-Review: The Contribution of Intermediate Phenotypes to GxE Effects on Disorders of Body Composition in the New OMICS Era" International Journal of Environmental Research and Public Health 14, no. 9: 1079. https://doi.org/10.3390/ijerph14091079