Biogenic and Synthetic Peptides with Oppositely Charged Amino Acids as Binding Sites for Mineralization
Abstract
:1. Introduction
- Do we find oppositely charged amino acids in other natural biomineralization proteins, and is this a characteristic feature of species- and/or mineral-specific biomineralization processes, especially with respect to CaCO3?
- Is there a general mode of action of peptides carrying oppositely charged amino acids to interact with materials other than the natural SiO2 or CaCO3 biominerals? In particular, how frequently do peptides identified by means of phage display and material screening approaches exhibit such properties?
- Is it possible to transfer features such as oppositely charged residues known from other natural mineral systems (exemplified for domains contributing to SiO2 formation here), to guide and control mineral deposition onto heterologous carrier templates such as tobacco mosaic virus (TMV)-based nanotubes and rings, thereby reducing the degrees of freedom of the peptides immobilized on TMV?
2. Materials and Methods
2.1. Sreening of Biomineralization Proteins
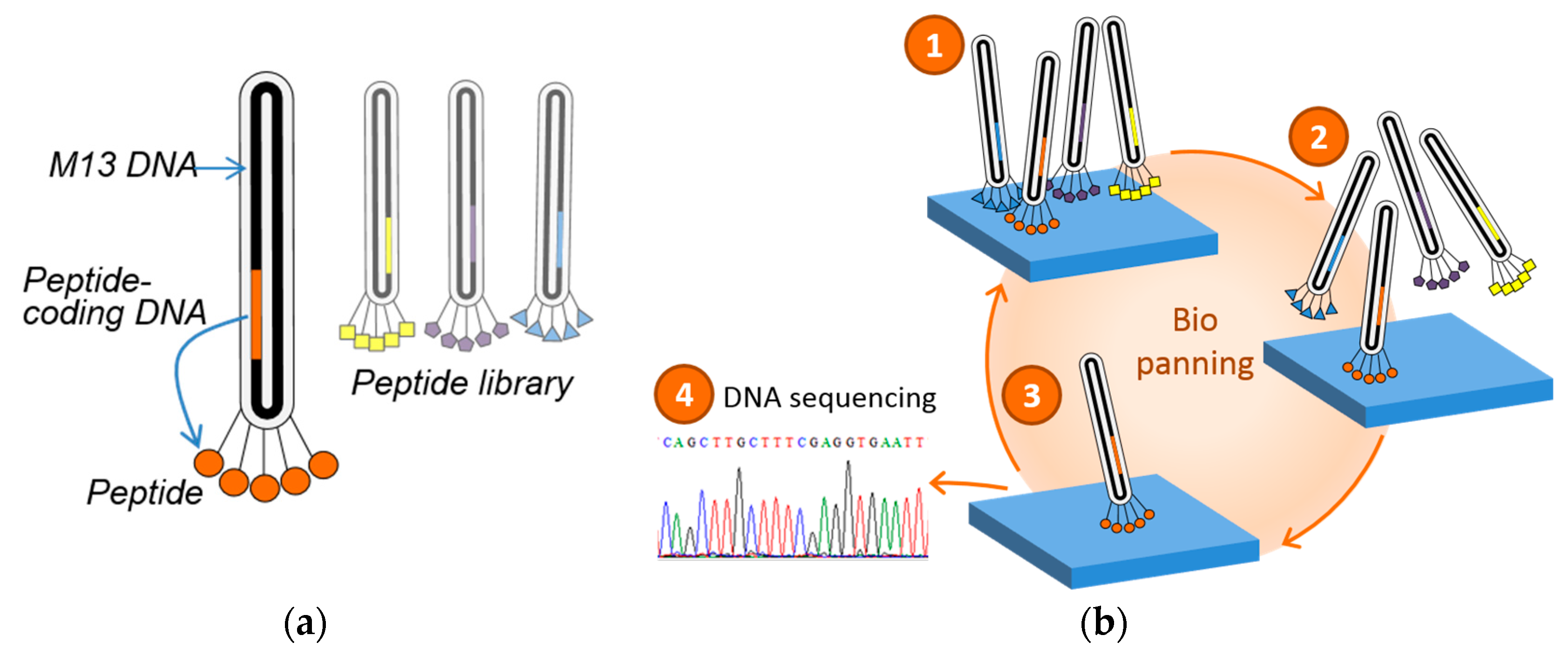
2.2. Phage Display
2.3. Re-Evaluation of Inorganic-Binding Peptides
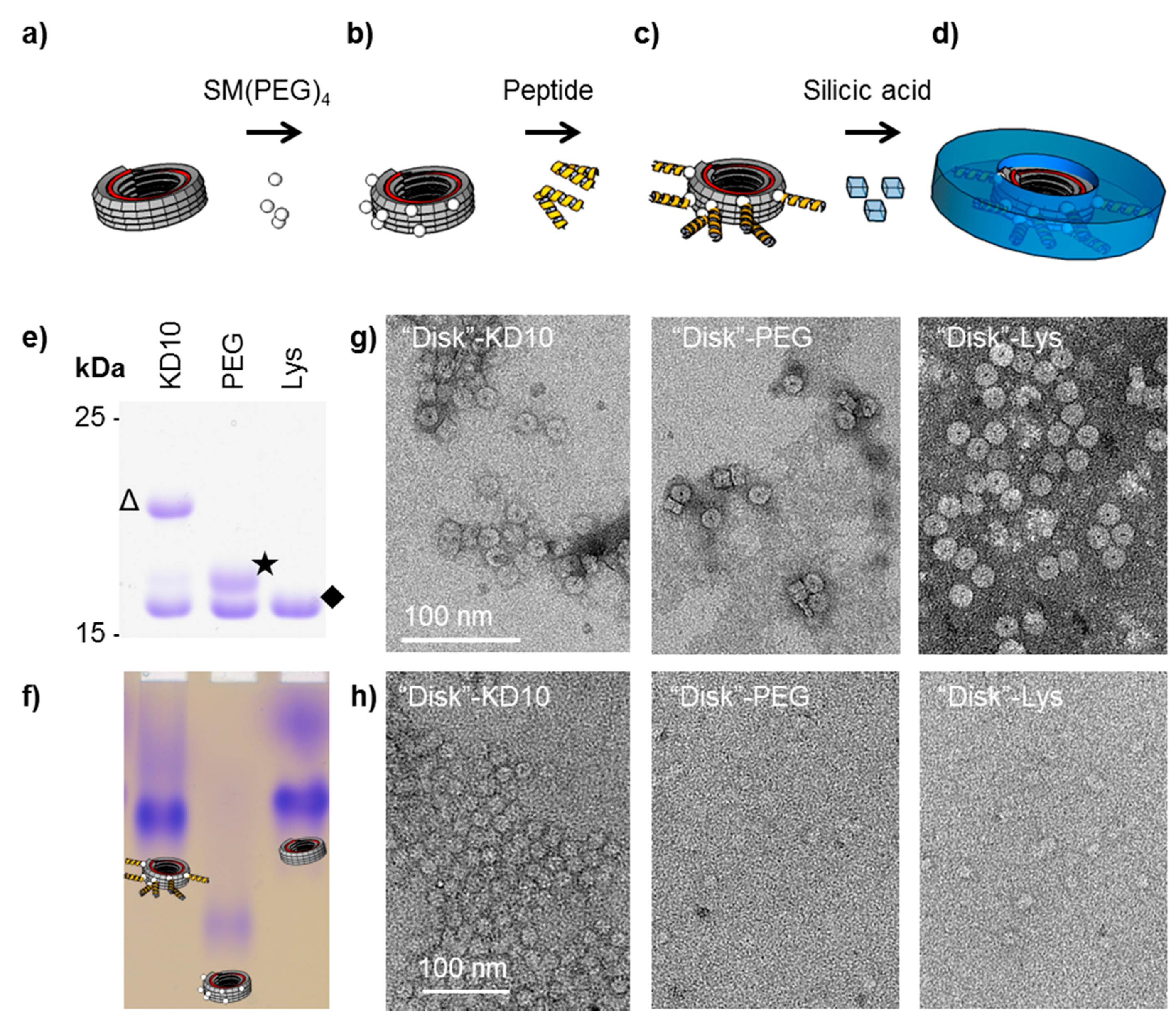
2.4. Silicification on TMV-Coupled Peptides
3. Results
3.1. Oppositely Charged Amino Acid Motifs in Biomineralization Proteins
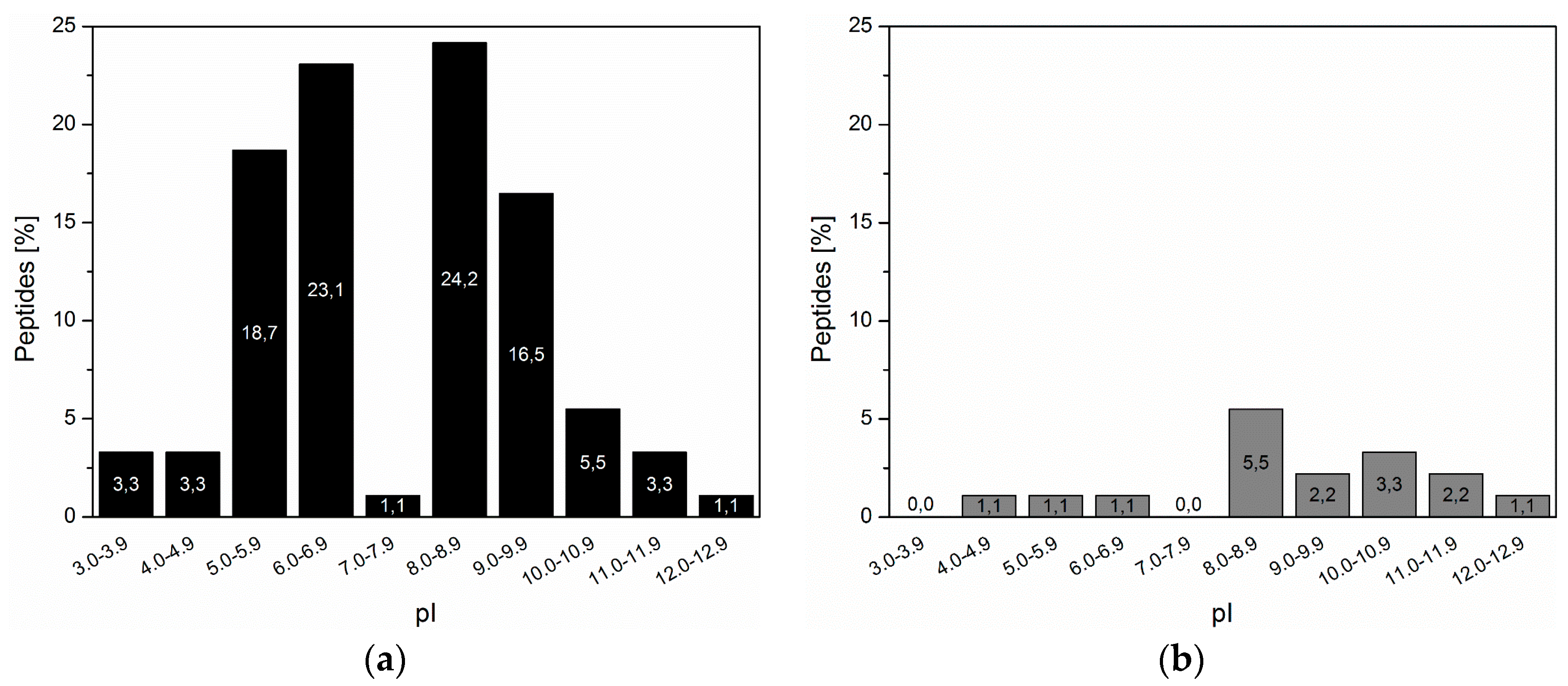
3.2. Inorganic Binding Peptides Identified by Phage Display
3.3. TMV-Coupled Bio-Inspired Peptides for Silicification
4. Discussion and Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Begum, G.; Goodwin, W.B.; deGlee, B.M.; Sandhage, K.H.; Kröger, N. Compartmentalisation of enzymes for cascade reactions through biomimetic layer-by-layer mineralization. J. Mater. Chem. 2015, 3, 5232–5240. [Google Scholar] [CrossRef]
- Brena, B.M.; Batista-Viera, F. Immobilization of Enzymes. In Immobilization of Enzymes and Cells; Guisán, J.M., Ed.; Humana Press: New York, NY, USA, 2006; Volume 22, pp. 15–30. [Google Scholar]
- Rothenstein, D.; Facey, S.J.; Ploss, M.; Hans, P.; Melcher, M.; Srot, V.; van Aken, P.A.; Hauer, B.; Bill, J. Mineralization of gold nanoparticles using tailored M13 phages. Bioinsp. Biomim. Nanobiomater. 2013, 2, 173–185. [Google Scholar] [CrossRef]
- Borg, S.; Rothenstein, D.; Bill, J.; Schüler, D. Generation of Multishell Magnetic Hybrid Nanoparticles by Encapsulation of Genetically Engineered and Fluorescent Bacterial Magnetosomes with ZnO and SiO2. Small 2015, 11, 4209–4217. [Google Scholar] [CrossRef] [PubMed]
- Rothenstein, D.; Shopova-Gospodinova, D.; Bakradze, G.; Jeurgens, L.P.H.; Bill, J. Generation of luminescence in biomineralized zirconia by zirconia-binding peptides. CrystEngComm 2015, 17, 1783–1790. [Google Scholar] [CrossRef]
- Atanasova, P.; Rothenstein, D.; Schneider, J.J.; Hoffmann, R.C.; Dilfer, S.; Eiben, S.; Wege, C.; Jeske, H.; Bill, J. Virus-Templated Synthesis of ZnO Nanostructures and Formation of Field-Effect Transistors. Adv. Mater. 2011, 23, 4918–4922. [Google Scholar] [CrossRef] [PubMed]
- Marin, F.; Luquet, G.; Marie, B.; Medakovic, D. Molluscan shell proteins: primary structure, origin, and evolution. Curr. Top. Dev. Biol. 2008, 80, 209–276. [Google Scholar] [PubMed]
- Marin, F.; Marie, B.; Hamada, S.B.; Ramos-Silva, P.; Le Roy, N.; Guichard, N.; Wolf, S.E.; Montagnani, C.; Joubert, C.; Piquemal, D. 'Shellome': Proteins involved in mollusk shell biomineralization-diversity, functions. Recent Adv. Pearl Res. 2013, 15, 149–166. [Google Scholar]
- Weiss, I.M.; Marin, F. The Role of Enzymes in Biomineralization Processes. In Biomineralization; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2008; pp. 71–126. [Google Scholar]
- Weber, E.; Pokroy, B. Intracrystalline inclusions within single crystalline hosts: from biomineralization to bio-inspired crystal growth. CrystEngComm 2015, 17, 5873–5883. [Google Scholar] [CrossRef]
- Marin, F.; Bundeleva, I.; Takeuchi, T.; Immel, F.; Medakovic, D. Organic matrices in metazoan calcium carbonate skeletons: Composition, functions, evolution. J. Struct. Biol. 2016, 196, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Ghatak, A.S.; Koch, M.; Guth, C.; Weiss, I.M. Peptide Induced Crystallization of Calcium Carbonate on Wrinkle Patterned Substrate: Implications for Chitin Formation in Molluscs. Int. J. Mol. Sci. 2013, 14, 11842–11860. [Google Scholar] [CrossRef] [PubMed]
- Weiss, I.M.; Lüke, F.; Eichner, N.; Guth, C.; Clausen-Schaumann, H. On the function of chitin synthase extracellular domains in biomineralization. J. Struct. Biol. 2013, 183, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Pohl, A.; Weiss, I.M. Real-time monitoring of calcium carbonate and cationic peptide deposition on carboxylate-SAM using a microfluidic SAW biosensor. Beilstein J. Nanotechnol. 2014, 5, 1823–1835. [Google Scholar] [CrossRef] [PubMed]
- Weiss, I.M.; Kaufmann, S.; Mann, K.; Fritz, M. Purification and Characterization of Perlucin and Perlustrin, Two New Proteins from the Shell of the Mollusc Haliotis laevigata. Biochem. Biophys. Res. Commun. 2000, 267, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.; Guth, C.; Weiss, I.M. GFP facilitates native purification of recombinant perlucin derivatives and delays the precipitation of calcium carbonate. PLoS ONE 2012, 7, e46653. [Google Scholar] [CrossRef]
- Weber, E.; Weiss, I.M.; Colfen, H.; Kellermeier, M. Recombinant perlucin derivatives influence the nucleation of calcium carbonate. CrystEngComm 2016, 18, 8439–8444. [Google Scholar] [CrossRef]
- Kamel, B.; Voolstra, C.; Medina, M. BioMine-DB: A database for metazoan biomineralization proteins. PeerJ Preprints 2016, 4, e1983v2. [Google Scholar]
- Rothenstein, D.; Claasen, B.; Omiecienski, B.; Lammel, P.; Bill, J. Isolation of ZnO-Binding 12-mer Peptides and Determination of Their Binding Epitopes by NMR Spectroscopy. J. Am. Chem. Soc. 2012, 134, 12547–12556. [Google Scholar] [CrossRef] [PubMed]
- Rodi, D.J.; Soares, A.S.; Makowski, L. Quantitative Assessment of Peptide Sequence Diversity in M13 Combinatorial Peptide Phage Display Libraries. J. Mol. Biol. 2002, 322, 1039–1052. [Google Scholar] [CrossRef]
- Baier, J.; Naumburg, T.; Blumenstein, N.J.; Jeurgens, L.P.H.; Welzel, U.; Do, T.A.; Pleiss, J.; Bill, J. Bio-inspired mineralization of zinc oxide in presence of ZnO-binding peptides. Biointerface Res. Appl. Chem. 2012, 2, 380–391. [Google Scholar]
- Slocik, J.M.; Stone, M.O.; Naik, R.R. Synthesis of gold nanoparticles using multifunctional peptides. Small 2005, 1, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Forbes, L.M.; Goodwin, A.P.; Cha, J.N. Tunable Size and Shape Control of Platinum Nanocrystals from a Single Peptide Sequence. Chem. Mater. 2010, 22, 6524–6528. [Google Scholar] [CrossRef]
- Hall Sedlak, R.; Hnilova, M.; Grosh, C.; Fong, H.; Baneyx, F.; Schwartz, D.; Sarikaya, M.; Tamerler, C.; Traxler, B. Engineered Escherichia coli Silver-Binding Periplasmic Protein That Promotes Silver Tolerance. Appl. Environ. Microbiol. 2012, 78, 2289–2296. [Google Scholar] [CrossRef] [PubMed]
- Ploss, M.; Facey, S.J.; Bruhn, C.; Zemel, L.; Hofmann, K.; Stark, R.W.; Albert, B.; Hauer, B. Selection of peptides binding to metallic borides by screening M13 phage display libraries. BMC Biotechnol. 2014, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Peters, E.A.; Schatz, P.J.; Johnson, S.S.; Dower, W.J. Membrane Insertion Defects Caused by Positive Charges in the Early Mature Region of Protein-Piii of Filamentous Phage-Fd Can Be Corrected Prla Suppressors. J. Bacteriol. 1994, 176, 4296–4305. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Eber, F.J.; Azucena, C.; Förste, A.; Walheim, S.; Schimmel, T.; Bittner, A.; Jeske, H.; Gliemann, H.; Eiben, S.; et al. Novel roles for well-known players: from tobacco mosaic virus pests to enzymatically active assemblies. Beilstein J. Nanotechnol. 2016, 7, 613–629. [Google Scholar] [CrossRef] [PubMed]
- van Rijn, P.; van Bezouwen, L.S.; Fischer, R.; Boekema, E.J.; Boker, A.; Commandeur, U. Virus-SiO2 and Virus-SiO2-Au Hybrid Particles with Tunable Morphology. Part. Part. Syst. Charact. 2015, 32, 43–47. [Google Scholar] [CrossRef]
- Steinmetz, N.F.; Shah, S.N.; Barclay, J.E.; Rallapalli, G.; Lomonossoff, G.P.; Evans, D.J. Virus-Templated Silica Nanoparticles. Small 2009, 5, 813–816. [Google Scholar] [CrossRef] [PubMed]
- Aljabali, A.A.; Shah, S.N.; Evans-Gowing, R.; Lomonossoff, G.P.; Evans, D.J. Chemically-coupled-peptide-promoted virus nanoparticle templated mineralization. Integr. Biol. 2011, 3, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Aljabali, A.A.A.; Barclay, J.E.; Cespedes, O.; Rashid, A.; Staniland, S.S.; Lomonossoff, G.P.; Evans, D.J. Charge Modified Cowpea Mosaic Virus Particles for Templated Mineralization. Adv. Funct. Mater. 2011, 21, 4137–4142. [Google Scholar] [CrossRef]
- Aljabali, A.A.A.; Lomonossoff, G.P.; Evans, D.J. CPMV-Polyelectrolyte-Templated Gold Nanoparticles. Biomacromolecules 2011, 12, 2723–2728. [Google Scholar] [CrossRef] [PubMed]
- Shenton, W.; Douglas, T.; Young, M.; Stubbs, G.; Mann, S. Inorganic-organic nanotube composites from template mineralization of tobacco mosaic virus. Adv. Mater. 1999, 11, 253. [Google Scholar] [CrossRef]
- Fowler, C.E.; Shenton, W.; Stubbs, G.; Mann, S. Tobacco mosaic virus liquid crystals as templates for the interior design of silica mesophases and nanoparticles. Adv. Mater. 2001, 13, 1266–1269. [Google Scholar] [CrossRef]
- Royston, E.; Lee, S.Y.; Culver, J.N.; Harris, M.T. Characterization of silica-coated tobacco mosaic virus. J. Colloid Interface Sci. 2006, 298, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Royston, E.S.; Brown, A.D.; Harris, M.T.; Culver, J.N. Preparation of silica stabilized Tobacco mosaic virus templates for the production of metal and layered nanoparticles. J. Colloid Interface Sci. 2009, 332, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Kadri, A.; Maiss, E.; Amsharov, N.; Bittner, A.M.; Balci, S.; Kern, K.; Jeske, H.; Wege, C. Engineered Tobacco mosaic virus mutants with distinct physical characteristics in planta and enhanced metallization properties. Virus Res. 2011, 157, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Mueller, A.; Eber, F.J.; Azucena, C.; Petershans, A.; Bittner, A.M.; Gliemann, H.; Jeske, H.; Wege, C. Inducible site-selective bottom-up assembly of virus-derived nanotube arrays on RNA-equipped wafers. ACS Nano 2011, 5, 4512–4520. [Google Scholar] [CrossRef] [PubMed]
- Eber, F.J.; Eiben, S.; Jeske, H.; Wege, C. Bottom-up-assembled nanostar colloids of gold cores and tubes derived from tobacco mosaic virus. Angew. Chem. 2013, 52, 7203–7207. [Google Scholar] [CrossRef] [PubMed]
- Eber, F.J.; Eiben, S.; Jeske, H.; Wege, C. RNA-controlled assembly of tobacco mosaic virus-derived complex structures: from nanoboomerangs to tetrapods. Nanoscale 2015, 7, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.; Eber, F.J.; Wenz, N.; Altintoprak, K.; Jeske, H.; Eiben, S.; Wege, C. Dynamic DNA-controlled "stop-and-go" assembly of well-defined protein domains on RNA-scaffolded TMV-like nanotubes. Nanoscale 2016, 8, 19853–19866. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Eber, F.; Nagarajan, A.S.; DiFranco, N.A.; Schmidt, N.; Wen, A.M.; Eiben, S.; Twyman, R.M.; Wege, C.; Steinmetz, N.F. The impact of aspect ratio on biodistribution and tumor homing of rigid soft-matter nanorods. Adv. Healthc. Mater. 2015, 4, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Mueller, A.; Degenhard, S.; Ruff, S.E.; Geiger, F.; Bittner, A.; Wege, C.; Krill, C., III. Enhancing the magnetoviscosity of ferrofluids by the addition of biological nanotubes. ACS Nano 2010, 4, 4531–4538. [Google Scholar] [CrossRef] [PubMed]
- Altintoprak, K.; Seidenstücker, A.; Krolla-Sidenstein, P.; Plettl, A.; Jeske, H.; Gliemann, H.; Wege, C. RNA-stabilized protein nanorings: High-precision adapters for biohybrid design. 2017; submitted. [Google Scholar]
- Kotzsch, A.; Pawolski, D.; Milentyev, A.; Shevchenko, A.; Scheffel, A.; Poulsen, N.; Kroger, N. Biochemical Composition and Assembly of Biosilica-associated Insoluble Organic Matrices from the Diatom Thalassiosira pseudonana. J. Biol. Chem. 2016, 291, 4982–4997. [Google Scholar] [CrossRef] [PubMed]
- Kroger, N.; Deutzmann, R.; Sumper, M. Polycationic peptides from diatom biosilica that direct silica nanosphere formation. Science 1999, 286, 1129–1132. [Google Scholar] [PubMed]
- Kroger, N.; Lorenz, S.; Brunner, E.; Sumper, M. Self-assembly of highly phosphorylated silaffins and their function in biosilica morphogenesis. Science 2002, 298, 584–586. [Google Scholar] [CrossRef] [PubMed]
- Sumper, M.; Hett, R.; Lehmann, G.; Wenzl, S. A code for lysine modifications of a silica biomineralizing silaffin protein. Angew. Chem. 2007, 46, 8405–8408. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, N.; Kroger, N. Silica morphogenesis by alternative processing of silaffins in the diatom Thalassiosira pseudonana. J. Biol. Chem. 2004, 279, 42993–42999. [Google Scholar] [CrossRef] [PubMed]
- Baio, J.E.; Zane, A.; Jaeger, V.; Roehrich, A.M.; Lutz, H.; Pfaendtner, J.; Drobny, G.P.; Weidner, T. Diatom Mimics: Directing the Formation of Biosilica Nanoparticles by Controlled Folding of Lysine-Leucine Peptides. J. Am. Chem. Soc. 2014, 136, 15134–15137. [Google Scholar] [CrossRef] [PubMed]
- Zane, A.C.; Michelet, C.; Roehrich, A.; Emani, P.S.; Drobny, G.P. Silica Morphogenesis by Lysine-Leucine Peptides with Hydrophobic Periodicity. Langmuir 2014, 30, 7152–7161. [Google Scholar] [CrossRef] [PubMed]
- Spinthaki, A.; Zerfaß, C.; Paulsen, H.; Hobe, S.; Demadis, K.D. Pleiotropic Role of Recombinant Silaffin-Like Cationic Polypeptide P5S3: Peptide-Induced Silicic Acid Stabilization, Silica Formation and Inhibition of Silica Dissolution. ChemistrySelect 2017, 2, 6–17. [Google Scholar] [CrossRef]
- Cha, J.N.; Shimizu, K.; Zhou, Y.; Christiansen, S.C.; Chmelka, B.F.; Stucky, G.D.; Morse, D.E. Silicatein filaments and subunits from a marine sponge direct the polymerization of silica and silicones in vitro. Proc. Natl. Acad. Sci. USA 1999, 96, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Schroder, H.C.; Wiens, M.; Schlossmacher, U.; Brandt, D.; Muller, W.E.G. Silicatein-Mediated Polycondensation of Orthosilicic Acid: Modeling of a Catalytic Mechanism Involving Ring Formation. Silicon-Neth 2012, 4, 33–38. [Google Scholar] [CrossRef]
- Weaver, J.C.; Morse, D.E. Molecular biology of demosponge axial filaments and their roles in biosilicification. Microsc. Res. Tech. 2003, 62, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Kuno, T.; Nonoyama, T.; Hirao, K.; Kato, K. Influence of the Charge Relay Effect on the Silanol Condensation Reaction as a Model for Silica Biomineralization. Langmuir 2011, 27, 13154–13158. [Google Scholar] [CrossRef] [PubMed]
- Murai, K.; Higuchi, M.; Kuno, T.; Kato, K. Silica Mineralization by a Peptide Template Having a High Charge Relay Effect. ChemPlusChem 2014, 79, 531–535. [Google Scholar] [CrossRef]
- Altintoprak, K.; Seidenstucker, A.; Welle, A.; Eiben, S.; Atanasova, P.; Stitz, N.; Plettl, A.; Bill, J.; Gliemann, H.; Jeske, H.; et al. Peptide-equipped tobacco mosaic virus templates for selective and controllable biomineral deposition. Beilstein J. Nanotechnol. 2015, 6, 1399–1412. [Google Scholar] [CrossRef] [PubMed]
- Butler, P.J. Self-assembly of tobacco mosaic virus: the role of an intermediate aggregate in generating both specificity and speed. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Pattanayek, R.; Stubbs, G. Structure of the U2 strain of tobacco mosaic virus refined at 3.5 A resolution using X-ray fiber diffraction. J. Mol. Biol. 1992, 228, 516–528. [Google Scholar] [CrossRef]
- Geiger, F.C.; Eber, F.J.; Eiben, S.; Mueller, A.; Jeske, H.; Spatz, J.P.; Wege, C. TMV nanorods with programmed longitudinal domains of differently addressable coat proteins. Nanoscale 2013, 5, 3808–3816. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.F.; DeYoreo, J.J.; Dove, P.M. Kinetics of Silica Nucleation on Carboxyl- and Amine-Terminated Surfaces: Insights for Biomineralization. J. Am. Chem. Soc. 2009, 131, 5244–5250. [Google Scholar] [CrossRef] [PubMed]
- Demadis, K.D.; Ketsetzi, A.; Pachis, K.; Ramos, V.M. Inhibitory Effects of Multicomponent, Phosphonate-Grafted, Zwitterionic Chitosan Biomacromolecules on Silicic Acid Condensation. Biomacromolecules 2008, 9, 3288–3293. [Google Scholar] [CrossRef] [PubMed]
- Demadis, K.D.; Pachis, K.; Ketsetzi, A.; Stathoulopoulou, A. Bioinspired control of colloidal silica in vitro by dual polymeric assemblies of zwitterionic phosphomethylated chitosan and polycations or polyanions. Adv. Colloid Interface Sci. 2009, 151, 33–48. [Google Scholar] [CrossRef] [PubMed]

| Species | Protein | Motifs | Sequences | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aa1 | pI | K | R | D | E | mol% Charged | aa3 | pI | mol% Charged | ||
| A. rigida | Chitin synthase, At-CS_E22, pI9 | 46 | 9.87 | 10 | 3 | 2 | 9 | 52.2 | 102 | 9.80 | 41.2 |
| H. laevigata | Perlucin C protein precursor | 42 | 7.88 | 0 | 5 | 5 | 0 | 33.3 | 240 | 7.02 | 25.4 |
| B. taurus | Extracellular Ca-sensing receptor | 40 | 8.47 | 4 | 3 | 2 | 4 | 32.5 | 1085 | 6.05 | 21.3 |
| R. norvegicus | Guanine nucleotide-binding protein | 26 | 10.60 | 5 | 5 | 2 | 5 | 65.4 | 1144 | 4.45 | 26 |
| R. norvegicus | Osteopontin | 50 | 4.57 | 5 | 2 | 8 | 6 | 50.0 | 317 | 4.12 | 27.2 |
| R. norvegicus | Plasma membrane Ca-transporting ATPase | 37 | 10.92 | 5 | 6 | 2 | 5 | 48.6 | 1181 | 5.86 | 25.7 |
| R. norvegicus | Collagen alpha-1(II) chain | 40 | 10.48 | 3 | 5 | 4 | 2 | 35.0 | 1419 | 8.44 | 19.1 |
| D. rerio | Exostosin-2 | 42 | 7.54 | 2 | 6 | 2 | 6 | 40.5 | 719 | 7.31 | 24.9 |
| P. fucata | Serine/threonine-protein kinase H1 | 46 | 11.16 | 7 | 7 | 3 | 3 | 45.7 | 415 | 10.22 | 31.6 |
| P. fucata | N16.5 matrix protein | 29 | 6.34 | 3 | 4 | 4 | 3 | 48.3 | 129 | 6.46 | 28.7 |
| P. fucata | Shematrin-5 | 34 | 3.77 | 0 | 5 | 12 | 1 | 52.9 | 278 | 7.59 | 23.4 |
| P. margaritifera | Calconectin | 30 | 4.75 | 4 | 2 | 6 | 1 | 43.3 | 92 | 6.65 | 41.3 |
| T. thermophila | Motor kinesin-like protein | 32 | 4.69 | 8 | 1 | 5 | 6 | 62.5 | 424 | 9.30 | 28.1 |
| Sequence | Negatively Charged AA | Positively Charged AA | Duplet | Net Charge pH 7 | pI1 |
|---|---|---|---|---|---|
| APSQPKDEVTAY | 2 | 1 | Yes | −1 | 4.37 |
| NPTLHQERMQDW | 2 | 1 | Yes | −1 | 5.32 |
| DLNYFTLSSKRE | 2 | 2 | Yes | 0 | 6.07 |
| ILSTQDLKAKSS | 1 | 2 | No | +1 | 8.59 |
| HYPTAKFHAERL | 1 | 2 | Yes | +1 | 8.60 |
| NLPKRDGWPLPW | 1 | 2 | Yes | +1 | 8.75 |
| YSLRADSRWMPS | 1 | 2 | No | +1 | 8.75 |
| FHEKQLSGGRFG | 1 | 2 | Yes | +1 | 8.76 |
| AIMGPRTVDRLP | 1 | 2 | Yes | +1 | 9.60 |
| IHSLQPEVNVRR | 1 | 2 | No | +1 | 9.61 |
| QHVYHPKPKASR | 0 | 3 | No | +3 | 10.29 |
| HLPQKKKPPHAI | 0 | 3 | No | +3 | 10.30 |
| HLPQKKKPPHAM | 0 | 3 | No | +3 | 10.30 |
| NKWPLAHSQKKR | 0 | 4 | No | +4 | 11.26 |
| KKRRRPSMYVPI | 0 | 5 | No | +5 | 11.73 |
| RKKTPASRRPMR | 0 | 6 | No | +6 | 12.48 |
| Sequence | Negatively Charged AA | Positively Charged AA | Duplet | Net Charge pH 7 | pI 1 |
|---|---|---|---|---|---|
| ERSWTLDSALSM | 2 | 1 | Yes | −1 | 4.37 |
| MKPDKAIRLDLL | 2 | 3 | Yes | +1 | 8.59 |
| HYPTAKFHAERL | 1 | 2 | Yes | +1 | 8.60 |
| HHTHRVDVHQTR | 1 | 2 | No | +1 | 9.62 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lemloh, M.-L.; Altintoprak, K.; Wege, C.; Weiss, I.M.; Rothenstein, D. Biogenic and Synthetic Peptides with Oppositely Charged Amino Acids as Binding Sites for Mineralization. Materials 2017, 10, 119. https://doi.org/10.3390/ma10020119
Lemloh M-L, Altintoprak K, Wege C, Weiss IM, Rothenstein D. Biogenic and Synthetic Peptides with Oppositely Charged Amino Acids as Binding Sites for Mineralization. Materials. 2017; 10(2):119. https://doi.org/10.3390/ma10020119
Chicago/Turabian StyleLemloh, Marie-Louise, Klara Altintoprak, Christina Wege, Ingrid M. Weiss, and Dirk Rothenstein. 2017. "Biogenic and Synthetic Peptides with Oppositely Charged Amino Acids as Binding Sites for Mineralization" Materials 10, no. 2: 119. https://doi.org/10.3390/ma10020119






