Single-Step Metal-Free Grafting of Cationic Polymer Brushes on Fluorescent Nanodiamonds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of FND
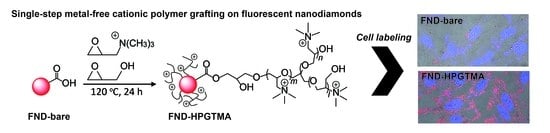
2.2. Synthesis of FND-PEGTMA, FND-HPGTMA, and FND-PEI
2.3. Characterization of FND Samples
2.4. Cell Culture, Labeling, Magnetically Modulated Fluorescence (MMF), and Confocal Fluorescence Microscopic Observation
2.5. Cell Viability Assay
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Krishnamoorthy, M.; Hakobyan, S.; Ramstedt, M.; Gautrot, J.E. Surface-initiated polymer brushes in the biomedical field: Applications in membrane science, biosensing, cell culture, regenerative medicine and antibacterial coatings. Chem. Rev. 2014, 114, 10976–11026. [Google Scholar] [CrossRef] [PubMed]
- Zoppe, J.O.; Ataman, N.C.; Mocny, P.; Wang, J.; Moraes, J.; Klok, H.A. Surface-Initiated Controlled Radical Polymerization: State-of-the-Art, Opportunities, and Challenges in Surface and Interface Engineering with Polymer Brushes. Chem. Rev. 2017, 117, 1105–1318. [Google Scholar] [CrossRef] [PubMed]
- Rosilo, H.; McKee, J.R.; Kontturi, E.; Koho, T.; Hytönen, V.P.; Ikkala, O.; Kostiainen, M.A. Cationic polymer brush-modified cellulose nanocrystals for high-affinity virus binding. Nanoscale 2014, 6, 11871–11881. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Lin, I.C.; Whittaker, M.R.; Minchin, R.F.; Monteiro, M.J.; Toth, I. Cellular Uptake of Densely Packed Polymer Coatings on Gold Nanoparticles. ACS Nano 2010, 4, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Yang, J.; Li, W.; Wang, W.; Liu, C.; Griffith, M.; Liu, W. Cationic polymer brush grafted-nanodiamond via atom transfer radical polymerization for enhanced gene delivery and bioimaging. J. Mater. Chem. 2011, 21, 7755–7764. [Google Scholar] [CrossRef]
- Matyjaszewski, K.; Xia, J. Atom transfer radical polymerization. Chem. Rev. 2001, 101, 2921–2990. [Google Scholar] [CrossRef] [PubMed]
- Lowe, A.B.; McCormick, C.L. Reversible addition-fragmentation chain transfer (RAFT) radical polymerization and the synthesis of water-soluble (co)polymers under homogeneous conditions in organic and aqueous media. Prog. Polym. Sci. 2007, 32, 283–351. [Google Scholar] [CrossRef]
- Dechy-Cabaret, O.; Martin-Vaca, B.; Bourissou, D. Controlled ring-opening polymerization of lactide and glycolide. Chem. Rev. 2004, 104, 6147–6176. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Huck, W.T.S. Hyperbranched polyglycidol on Si/SiO2 surfaces via surface-initiated polymerization. Macromolecules 2003, 36, 5088–5093. [Google Scholar] [CrossRef]
- Zhou, L.; Gao, C.; Xu, W.; Wang, X.; Xu, Y. Enhanced Biocompatibility and Biostability of CdTe Quantum Dots by Facile Surface-Initiated Dendritic Polymerization. Biomacromolecules 2009, 10, 1865–1874. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhou, Y.; Yang, S.; Ding, B. Growing hyperbranched polyglycerols on magnetic nanoparticles to resist nonspecific adsorption of proteins. Colloids Surf. B Biointerfaces 2008, 67, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Takimoto, T.; Ito, M.; Kitagawa, N.; Kimura, T.; Komatsu, N. Chromatographic Separation of Highly Soluble Diamond Nanoparticles Prepared by Polyglycerol Grafting. Angew. Chem. Int. Ed. 2011, 50, 1388–1392. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.J.; Kang, M.W.; Chang, H.C.; Chen, K.M.; Yu, Y.C. Bright Fluorescent Nanodiamonds: No Photobleaching and Low Cytotoxicity. J. Am. Chem. Soc. 2005, 127, 17604–17605. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.C.; Lee, H.Y.; Chen, K.; Lim, T.S.; Wu, H.Y.; Lin, P.K.; Wei, P.K.; Tsao, P.H.; Chang, H.C.; Fann, W. Characterization and application of single fluorescent nanodiamonds as cellular biomarkers. Proc. Natl. Acad. Sci. USA 2007, 104, 727–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohan, N.; Chen, C.S.; Hsieh, H.H.; Wu, Y.C.; Chang, H.C. In Vivo Imaging and Toxicity Assessments of Fluorescent Nanodiamonds in Caenorhabditis elegans. Nano Lett. 2010, 10, 3692–3699. [Google Scholar] [CrossRef] [PubMed]
- Hui, Y.Y.; Hsiao, W.W.W.; Haziza, S.; Simonneau, M.; Treussart, F.; Chang, H.C. Single particle tracking of fluorescent nanodiamonds in cells and organisms. Curr. Opin. Solid State Mater. Sci. 2017, 21, 35–42. [Google Scholar] [CrossRef]
- Hsiao, W.W.; Hui, Y.Y.; Tsai, P.; Chang, H. Fluorescent Nanodiamond: A Versatile Tool for Long-Term Cell Tracking, Super-Resolution Imaging, and Nanoscale Temperature Sensing. Acc. Chem. Res. 2016, 49, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Su, L.J.; Wu, M.S.; Hui, Y.Y.; Chang, B.M.; Pan, L.; Hsu, P.C.; Chen, Y.T.; Ho, H.N.; Huang, Y.H.; Ling, T.Y.; et al. Fluorescent nanodiamonds enable quantitative tracking of human mesenchymal stem cells in miniature pigs. Sci. Rep. 2017, 7, 45607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sotoma, S.; Akagi, K.; Hosokawa, S.; Igarashi, R.; Tochio, H.; Harada, Y.; Shirakawa, M. Comprehensive and quantitative analysis for controlling the physical/chemical states and particle properties of nanodiamonds for biological applications. RSC Adv. 2015, 5, 13818–13827. [Google Scholar] [CrossRef] [Green Version]
- Neburkova, J.; Vavra, J.; Cigler, P. Coating nanodiamonds with biocompatible shells for applications in biology and medicine. Curr. Opin. Solid State Mater. Sci. 2017, 21, 43–53. [Google Scholar] [CrossRef]
- Chang, Y.R.; Lee, H.Y.; Chen, K.; Chang, C.C.; Tsai, D.S.; Fu, C.C.; Lim, T.S.; Tzeng, Y.K.; Fang, C.Y.; Han, C.C.; et al. Mass production and dynamic imaging of fluorescent nanodiamonds. Nat. Nanotechnol. 2008, 3, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.K.; Bumb, A.; Wu, X.; Sochacki, K.A.; Kellman, P.; Brechbiel, M.W.; Neuman, K.C. Wide-field in vivo background free imaging by selective magnetic modulation of nanodiamond fluorescence. Biomed. Opt. Express 2014, 5, 1190. [Google Scholar] [CrossRef] [PubMed]
- Sotoma, S.; Shirakawa, M. Monodispersed Colloidal Solutions of Surface-modified Detonation-synthesized Nanodiamonds and Their Aggregation Resistance. Chem. Lett. 2016, 45, 697–699. [Google Scholar] [CrossRef]
- Wang, L.; Neoh, K.G.; Kang, E.T.; Shuter, B.; Wang, S.C. Superparamagnetic Hyperbranched Polyglycerol-Grafted Fe3O4 Nanoparticles as a Novel Magnetic Resonance Imaging Contrast Agent: An In Vitro Assessment. Adv. Funct. Mater. 2009, 19, 2615–2622. [Google Scholar] [CrossRef]
- Zhou, L.; Gao, C.; Xu, W. Robust Fe3O4/SiO2-Pt/Au/Pd magnetic nanocatalysts with multifunctional hyperbranched polyglycerol amplifiers. Langmuir 2010, 26, 11217–11225. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Guo, Z.; Feng, R.; Zhang, Y.; Xue, W.; Liu, Z. Hyperbranched polyglycerol conjugated fluorescent carbon dots with improved in vitro toxicity and red blood cell compatibility for bioimaging. RSC Adv. 2017, 7, 4975–4982. [Google Scholar] [CrossRef] [Green Version]
- Das, P.; Jana, N.R. Highly colloidally stable hyperbranched polyglycerol grafted red fluorescent silicon nanoparticle as bioimaging probe. ACS Appl. Mater. Interfaces 2014, 6, 4301–4309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Q.; Chen, M.; Lam, R.; Xu, X.; Osawa, E.; Ho, D. Polymer-Functionalized Nanodiamond Platforms as Vehicles for Gene Delivery. ACS Nano 2009, 3, 2609–2616. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sotoma, S.; Hsieh, F.-J.; Chang, H.-C. Single-Step Metal-Free Grafting of Cationic Polymer Brushes on Fluorescent Nanodiamonds. Materials 2018, 11, 1479. https://doi.org/10.3390/ma11081479
Sotoma S, Hsieh F-J, Chang H-C. Single-Step Metal-Free Grafting of Cationic Polymer Brushes on Fluorescent Nanodiamonds. Materials. 2018; 11(8):1479. https://doi.org/10.3390/ma11081479
Chicago/Turabian StyleSotoma, Shingo, Feng-Jen Hsieh, and Huan-Cheng Chang. 2018. "Single-Step Metal-Free Grafting of Cationic Polymer Brushes on Fluorescent Nanodiamonds" Materials 11, no. 8: 1479. https://doi.org/10.3390/ma11081479