Characterization of Biomaterials by Soft X-Ray Spectromicroscopy
Abstract
:1. Introduction
2. Spectromicroscopy Background
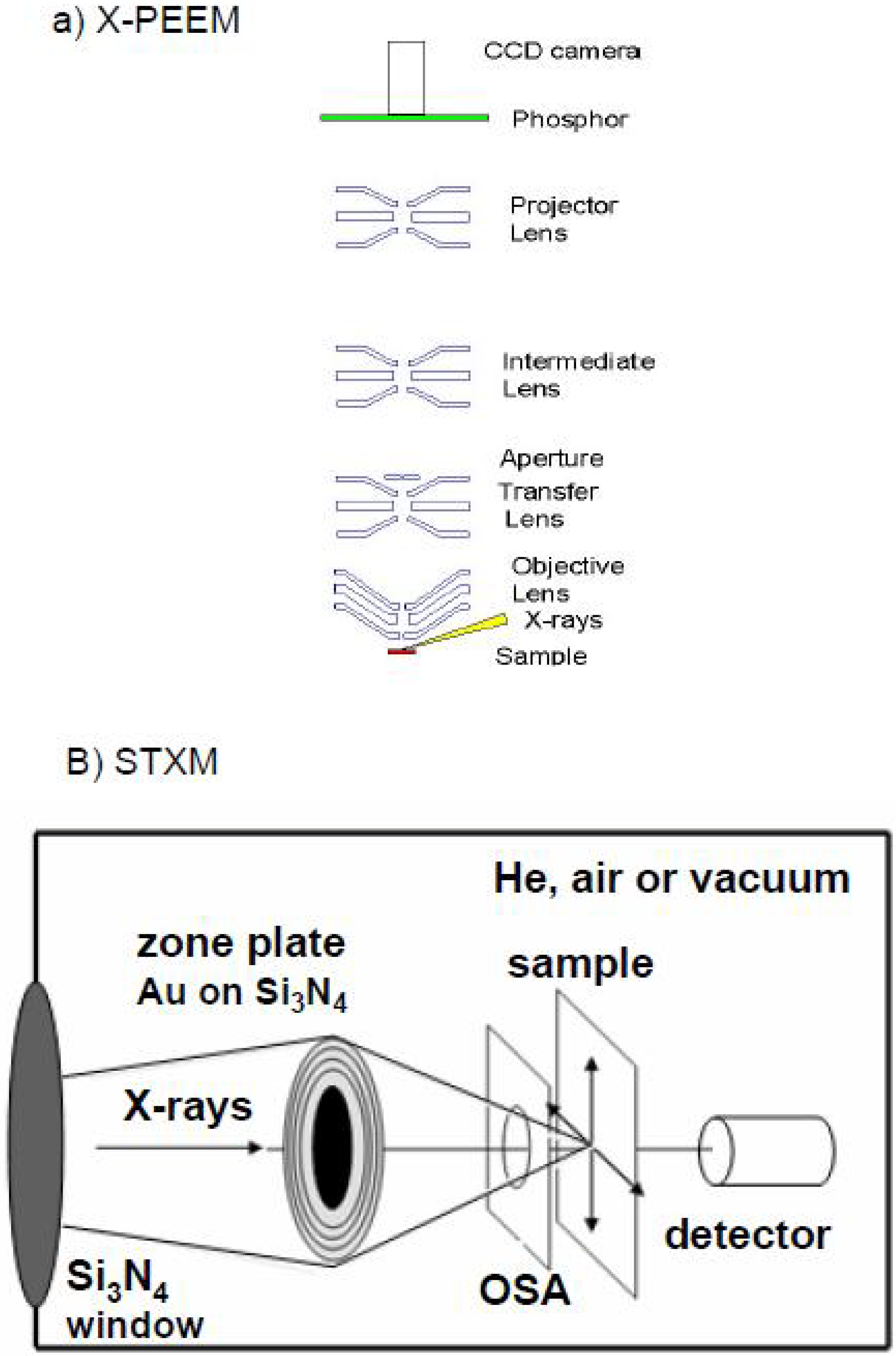
2.1. X-ray Photoemission Electron Microscopy (X-PEEM)
2.2. Scanning Transmission X-ray Microscopy (STXM)
2.3. Data Analysis
3. Results and Discussion
3.1. Spectromicroscopy Applications
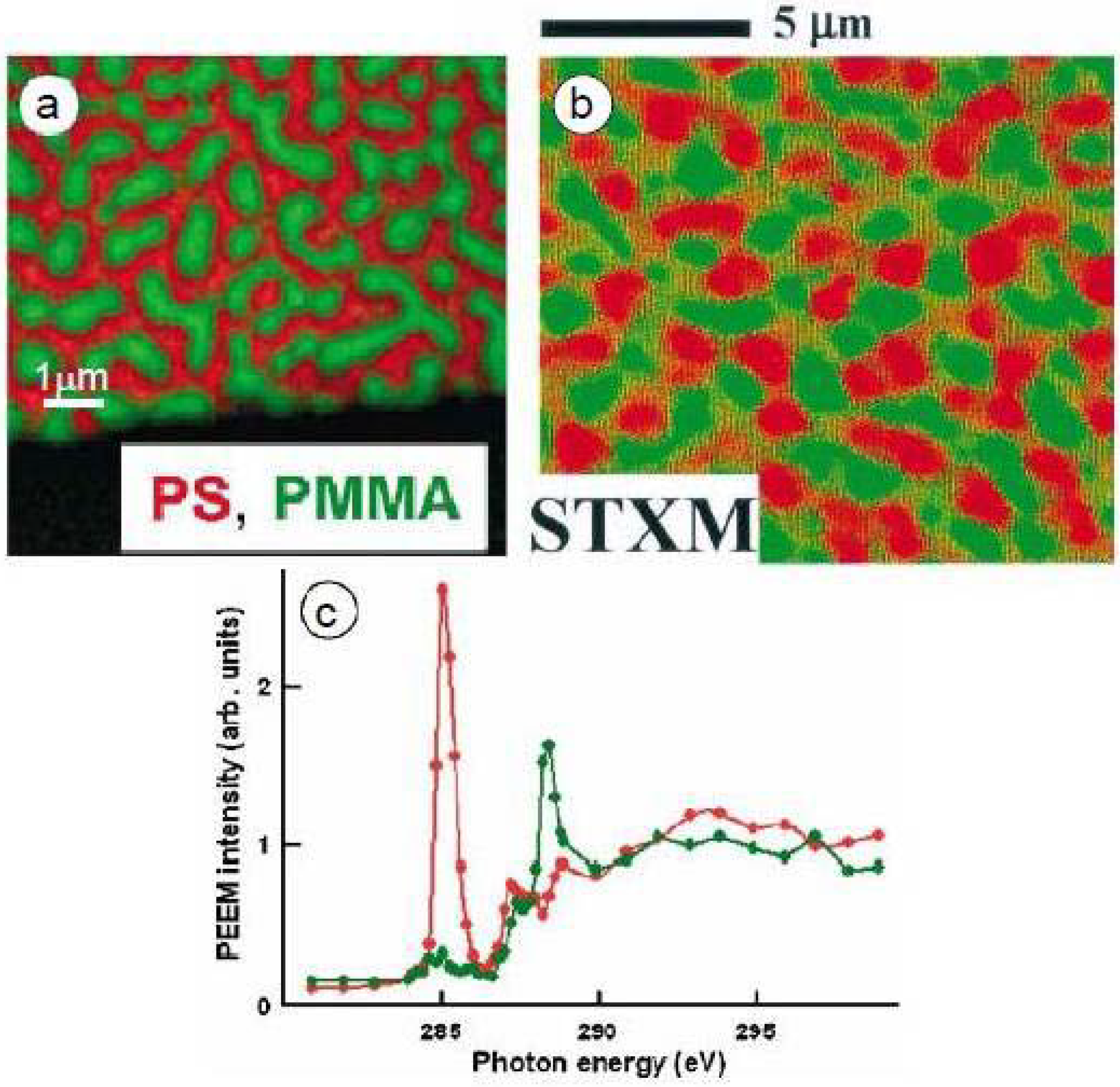
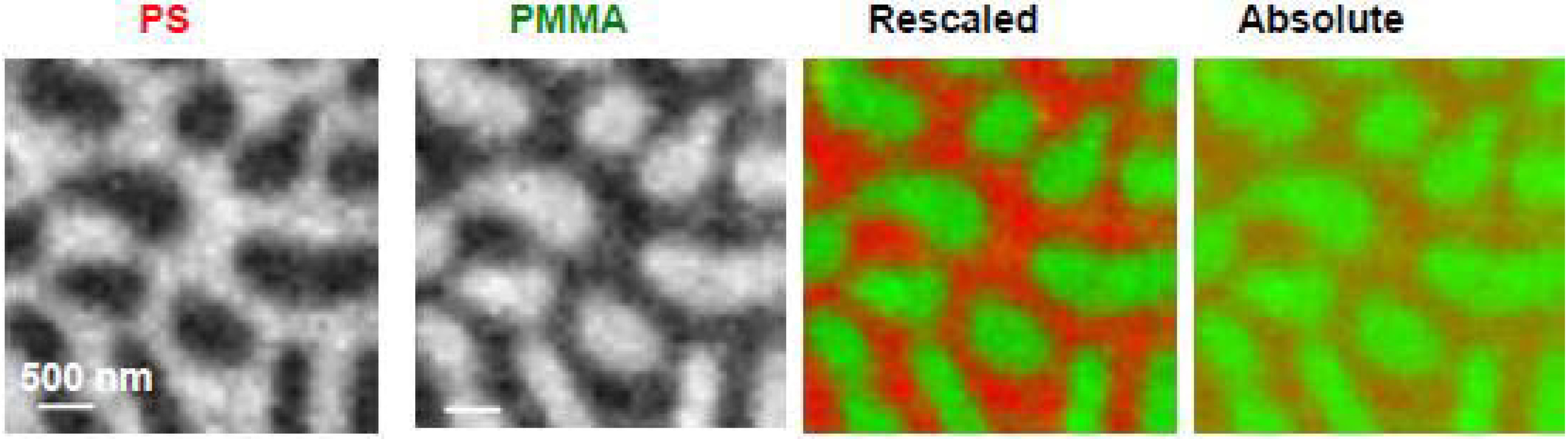
3.1.1. Polystyrene-Poly(methyl methacrylate) (PS-PMMA)
3.1.1.1. Optimization of PS-PMMA blend surface
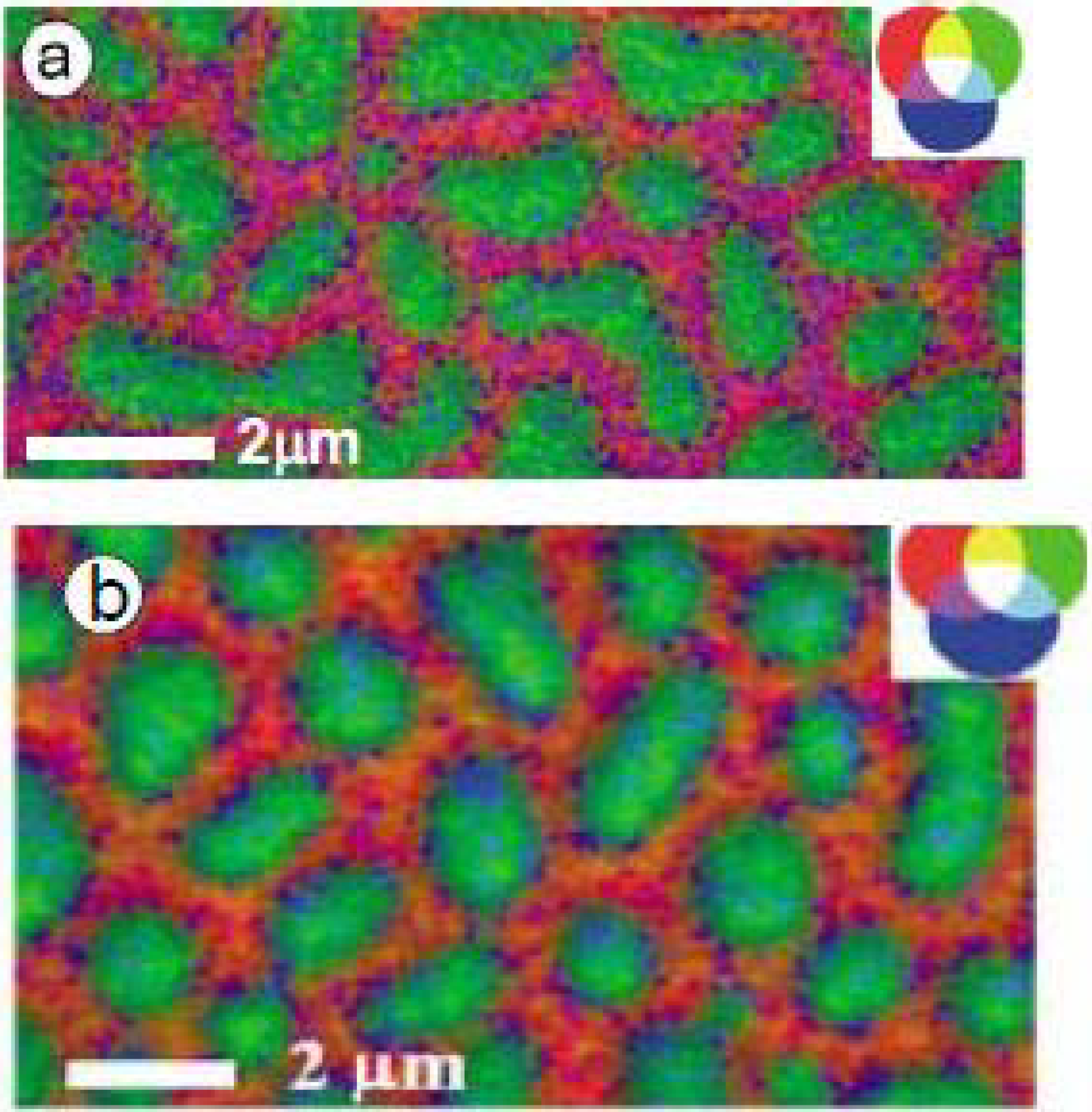
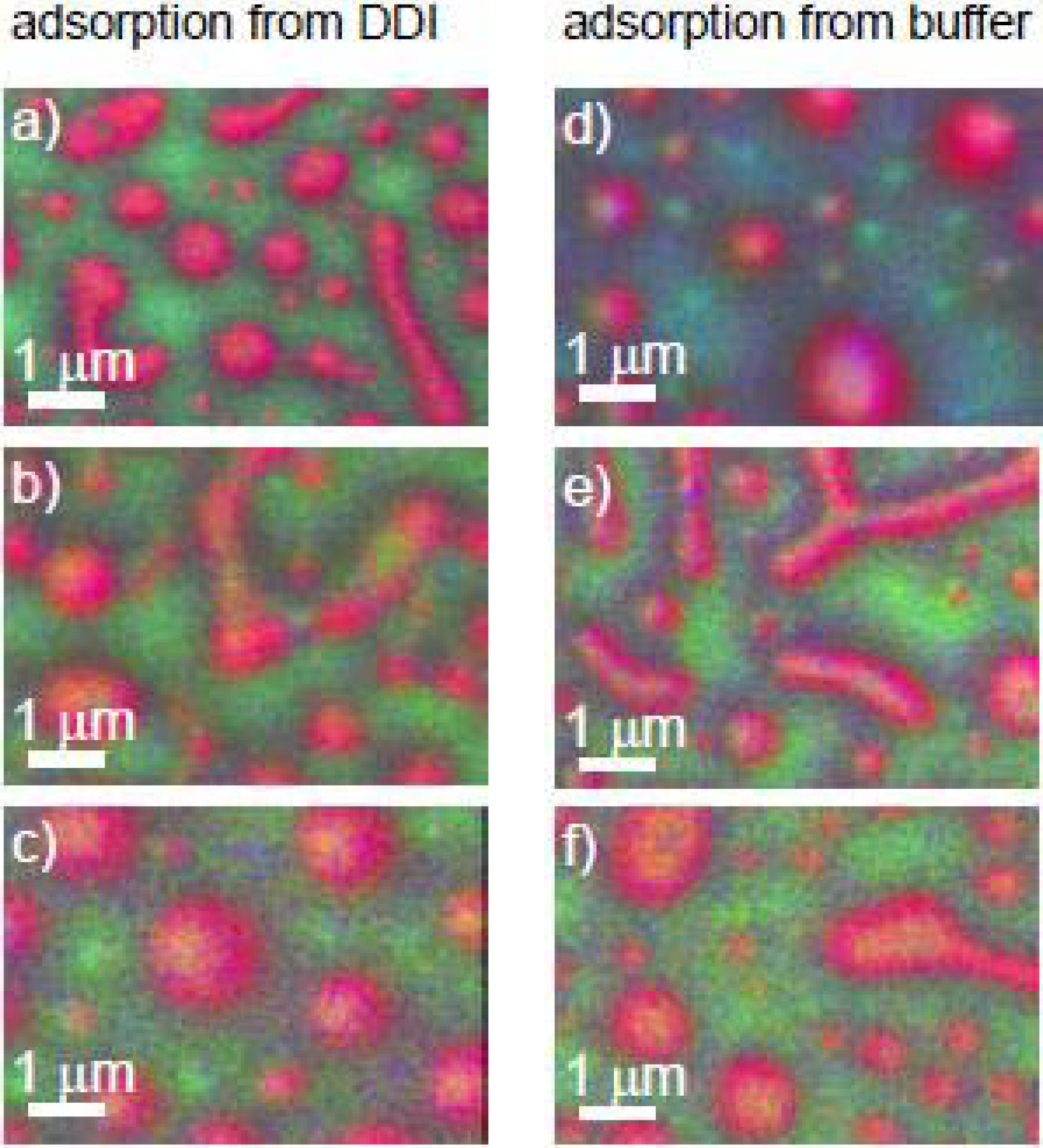
3.1.1.2. Fibrinogen (Fg) Adsorption to PS-PMMA—Buffer vs. Water
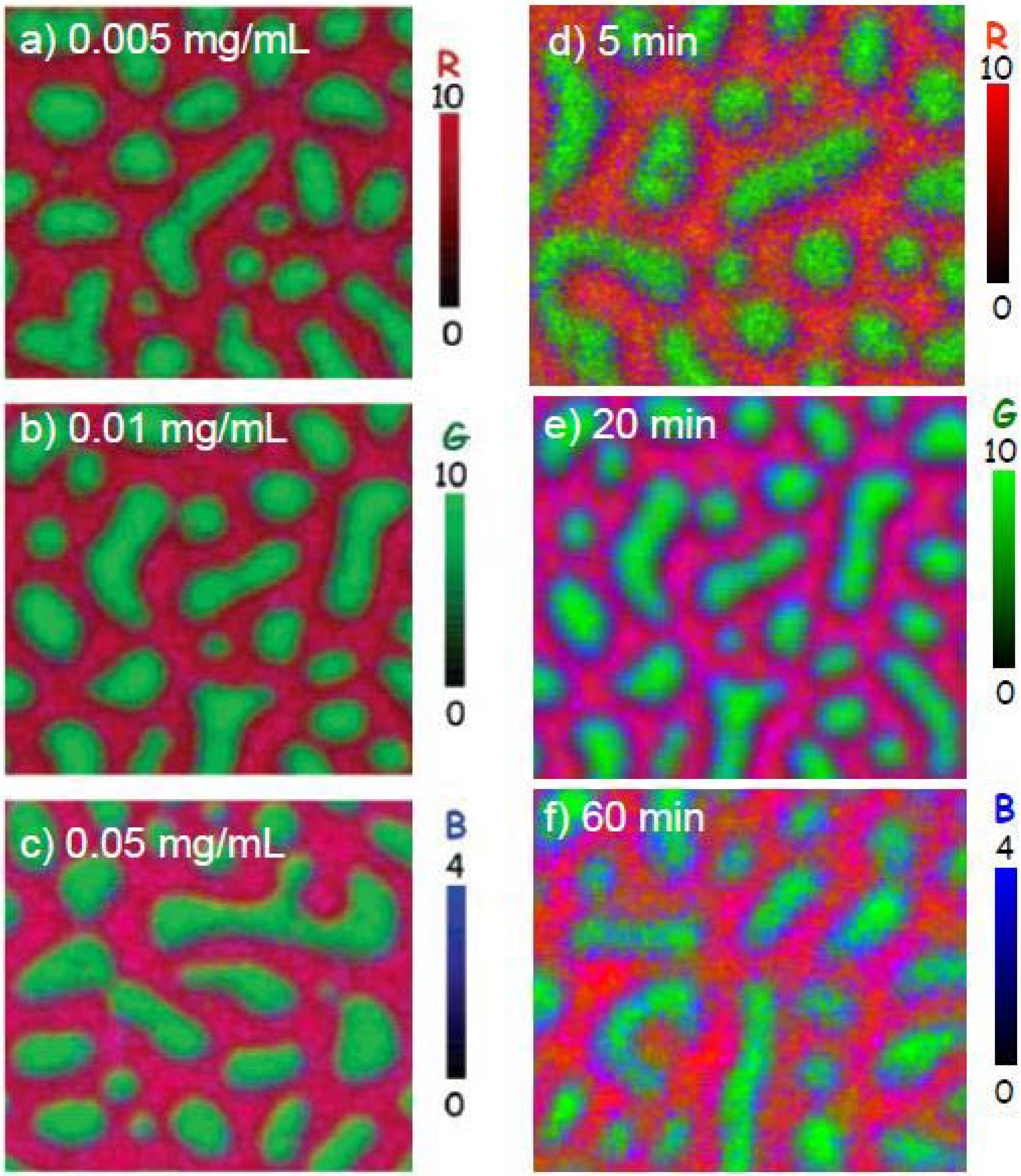
3.1.1.3. HSA adsorption to PS-PMMA

3.1.1.4. Protein-peptide competitive adsorption
3.1.2. Biodegradable or Biocompatible Polymers
3.1.2.1. HSA adsorption to polystyrene-polylacide (PS-PLA)
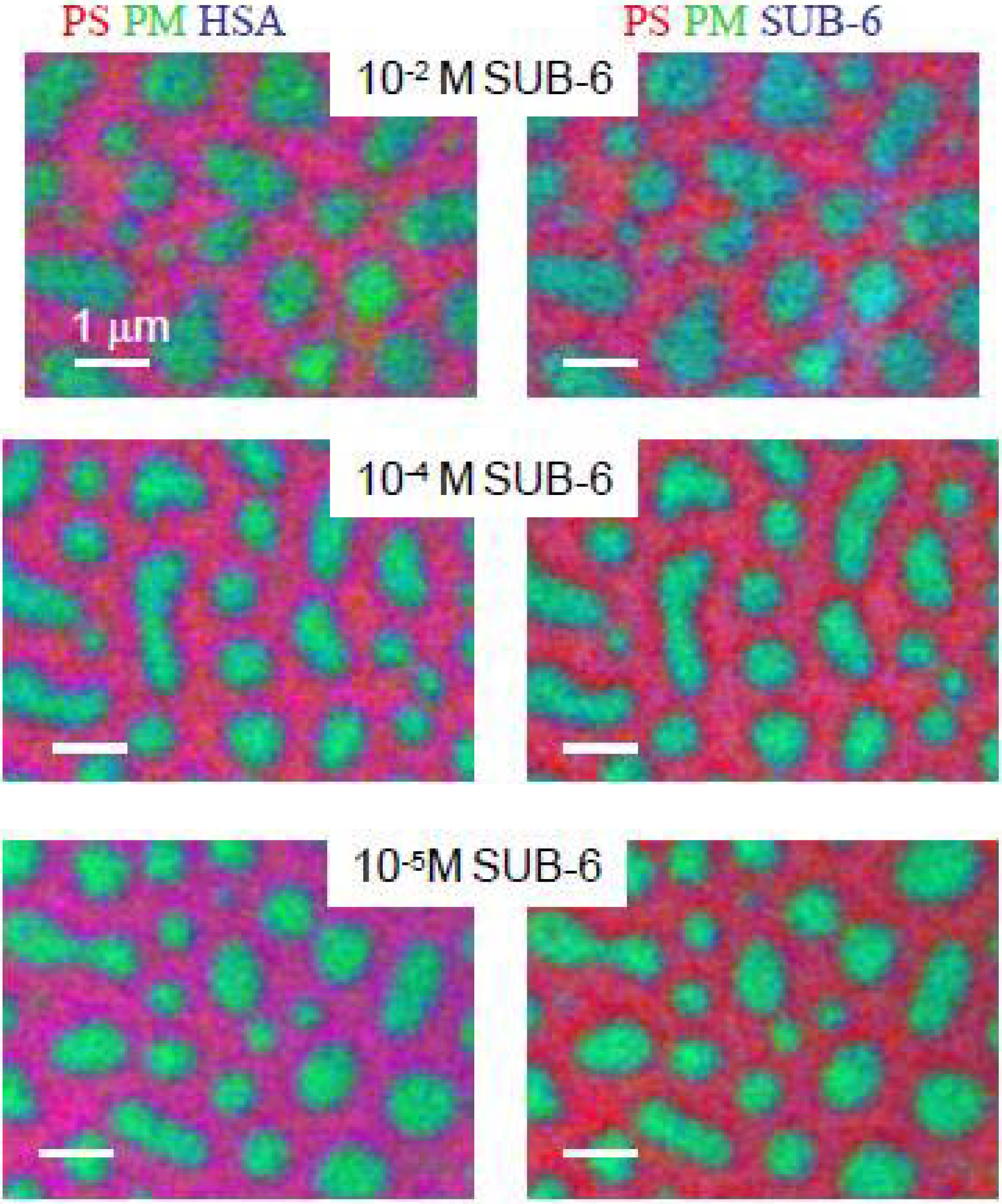
3.1.2.2. HSA adsorption to PMMA-chitosan blends
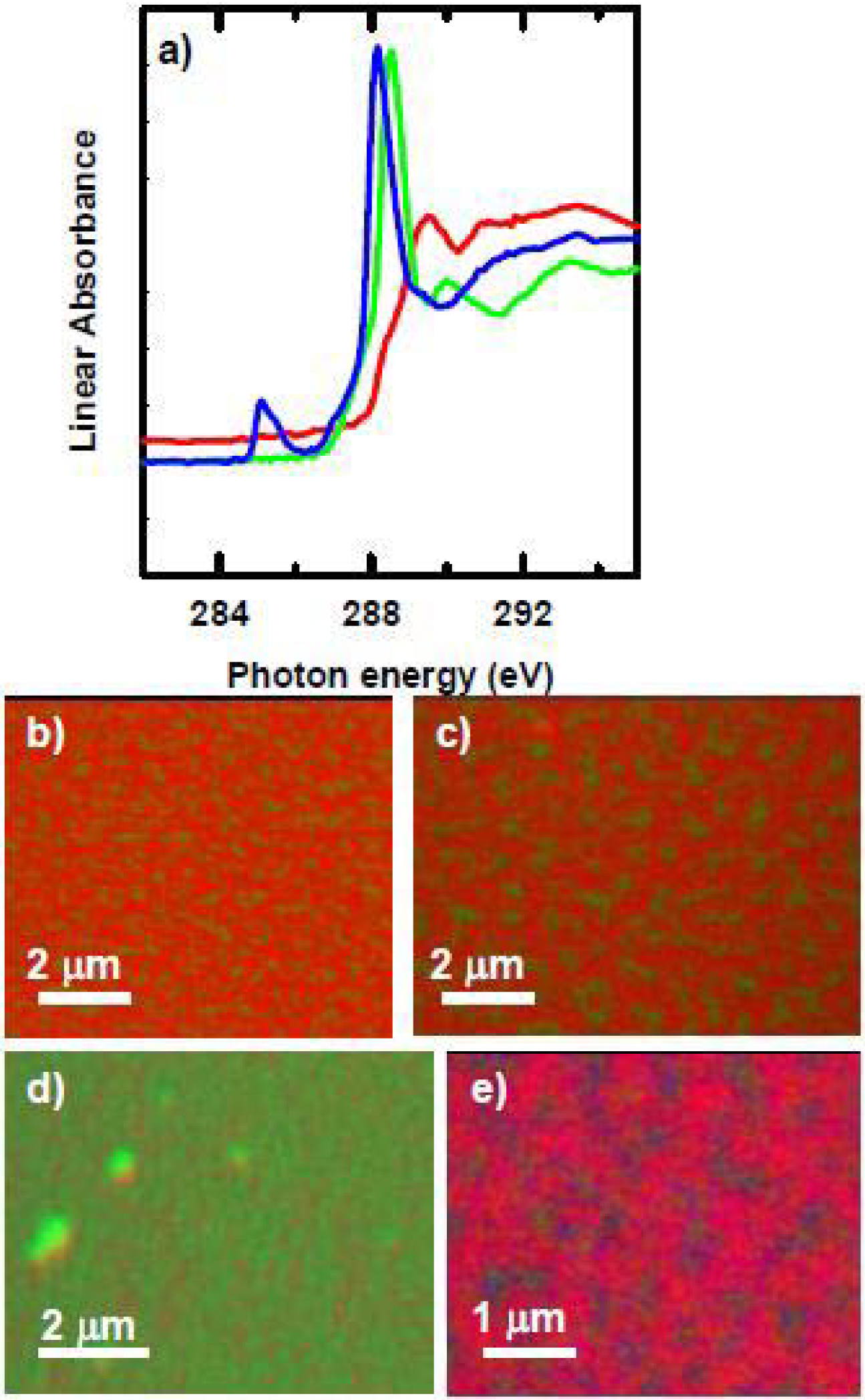
3.1.2.3. HSA adsorption to PEO-PETA blends
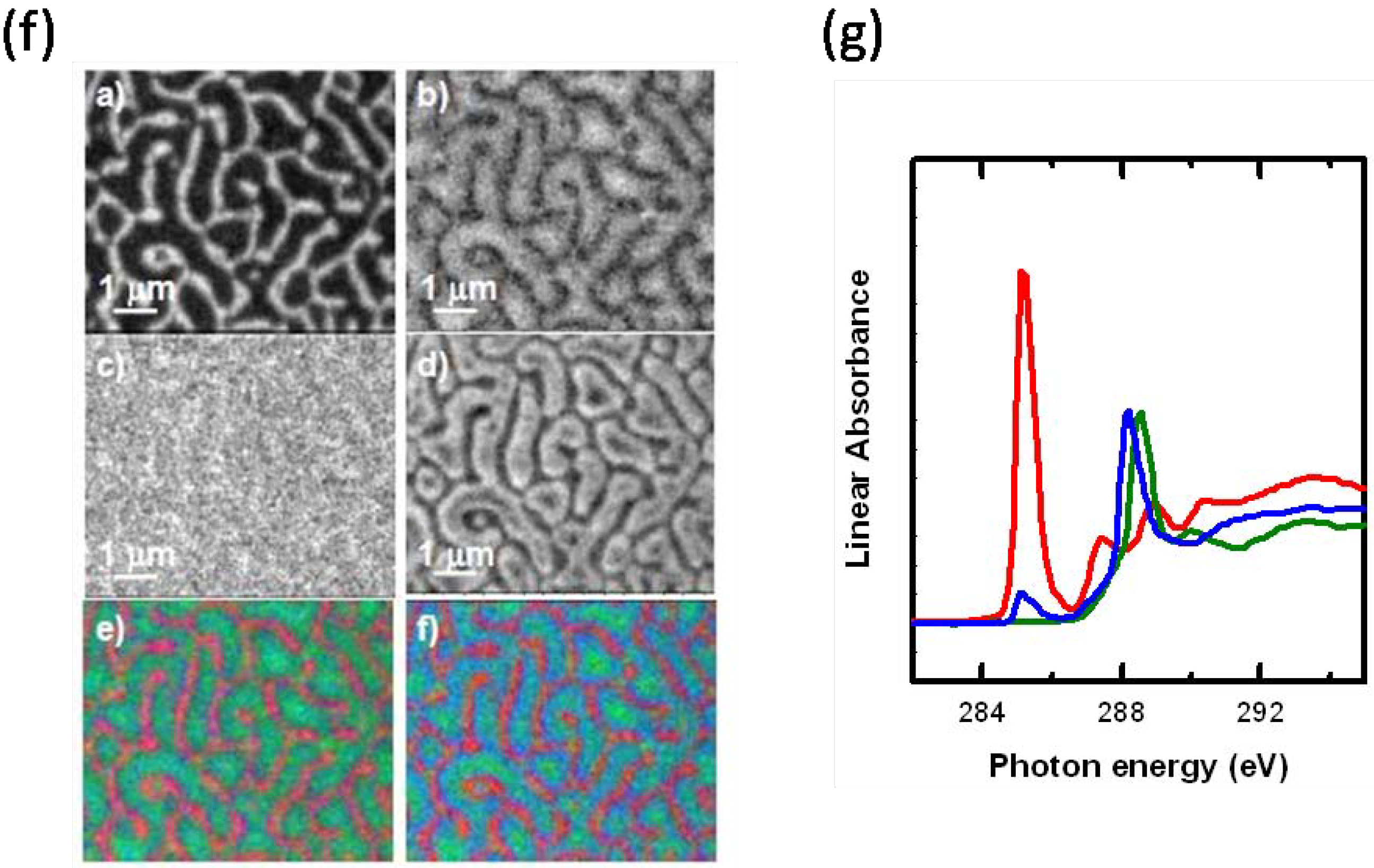
3.1.2.4. Fluoropolymers for stent coating applications (Substrate characterization)
3.1.3. Protein adsorption under hydrated conditions
3.1.3.1. Fg adsorption to poly(styrenecoacrylonitrile) (SAN) and poly-isocyanate poly-addition product (PIPA) nanoparticles embedded in polyurethane

3.1.3.2. HSA adsorption to PS/PMMA

3.1.3.3. Electrolyte-induced deswelling behavior of pH-responsive microgels
4. Summary and Conclusions
Acknowledgements
References and Notes
- Castner, D.G.; Ratner, B.D. Biomedical surface science: Foundations to frontiers. Surface Sci. 2002, 500, 28–60. [Google Scholar] [CrossRef]
- Elwing, H.; Welin, S.; Askendal, A.; Nilsson, U.; Lundstrom, I. A wettability gradient method for studies of macromolecular interactions at the liquid/solid interface. J. Colloid Interface Sci. 1987, 119, 203–210. [Google Scholar] [CrossRef]
- Ekblad, T.; Andersson, O.; Tai, F.I.; Ederth, T.; Liedberg, B. Lateral control of protein adsorption on charged polymer gradients. Langmuir 2009, 25, 3755–3762. [Google Scholar]
- Chittur, K.K. FTIR/ATR for protein adsorption to biomaterial surfaces. Biomaterials 1998, 19, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Green, R.J.; Frazier, R.A.; Shakesheff, K.M.; Davies, M.C.; Roberts, C.J.; Tendler, S.J.B. Surface plasmon resonance analysis of dynamic biological interactions with biomaterials. Biomaterials 2000, 21, 1823–1835. [Google Scholar] [CrossRef] [PubMed]
- Sigal, G.B.; Mrksich, M.; Whitesides, G.M. Effect of surface wettability on the adsorption of proteins and detergents. J. Am. Chem. Soc. 1998, 120, 3464–3473. [Google Scholar] [CrossRef]
- Hook, F.; Kasemo, B.; Nylander, T.; Fant, C.; Sott, K.; Elwing, H. Variations in coupled water, viscoelastic properties, and film thickness of a Mefp-1 protein film during adsorption and cross-linking: a quartz crystal microbalance with dissipation monitoring, ellipsometry, and surface plasmon resonance study. Anal. Chem. 2001, 73, 5796–5804. [Google Scholar] [CrossRef] [PubMed]
- Elwing, H. Protein absorption and ellipsometry in biomaterial research. Biomaterials 1998, 19, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Hook, F.; Rodahl, M.; Brzezinski, P.; Kasemo, B. Energy dissipation kinetics for protein and antibody−antigen adsorption under shear oscillation on a quartz crystal microbalance. Langmuir 1998, 14, 729–734. [Google Scholar] [CrossRef]
- Archambault, J.G.; Brash, J.L. Protein repellent polyurethane-urea surfaces by chemical grafting of hydroxyl-terminated poly(ethylene oxide): effects of protein size and charge. Colloid. Surface. B 2004, 33, 111–120. [Google Scholar] [CrossRef]
- Siedlecki, C.A.; Marchant, R.E. Atomic force microscopy for characterization of the biomaterial interface. Biomaterials 1998, 19, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Ta, T.C.; McDermott, M.T. Mapping interfacial chemistry induced variations in protein adsorption with scanning force microscopy. Anal Chem. 2000, 72, 2627–2634. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.; Sengonul, M.; Latour, R.; Kohn, J.; Libera, M. Selective protein adsorption on a phase-separated solvent-cast polymer blend. Langmuir 2006, 22, 6286–6292. [Google Scholar] [CrossRef] [PubMed]
- Bauer, E. Photoelectron spectromicroscopy: present and future. J. Electron Spectrosc. Relat. Phenom. 2001, 975, 114–116. [Google Scholar]
- Bauer, E. Photoelectron microscopy. J. Phys. Condens. Matter 2001, 13, 11391. [Google Scholar] [CrossRef]
- Kirz, J.; Jacobson, C.; Howells, M.Q. Soft x-ray microscopes and their biological applications. Rev. Biophys. 1995, 28, 33. [Google Scholar] [CrossRef]
- Ade, H. Experimental Methods in the Physical Sciences; Samson, J.A.R., Ederer, D.L., Eds.; Academic Press: San Diego, CA, USA, 1998; Volume 32, p. 225. [Google Scholar]
- Urquhart, S.G.; Ade, H. Chemical Applications of Synchrotron Radiation; Sham, T.K., Ed.; World Scientific: Singapore, 2002; p. 285. [Google Scholar]
- Ade, H.; Hitchcock, A.P. NEXAFS microscopy and resonant scattering: Composition and orientation probed in real and reciprocal space. Polymer 2008, 49, 643–675. [Google Scholar] [CrossRef]
- Sengonul, M.; Sousa, A.; Libera, M. Selective adsorption of surface-modified ferritin on a phase-separated polymer blend. Colloid. Surface B 2009, 73, 152–155. [Google Scholar] [CrossRef]
- Seo, J.H.; Matsuno, R.; Takai, M.; Ishihara, K. Cell adhesion on phase-separated surface of block copolymer composed of poly-(2-methacryloyloxyethyl phosphorylcholine) and poly(dimethylsiloxane). Biomaterials 2009, 30, 5330–5340. [Google Scholar] [CrossRef] [PubMed]
- Denisov, V.P.; Jonsson, B.H.; Halle, B. Hydration of denatured and molten globule proteins. Nat. Struct. Biol. 1999, 6, 253. [Google Scholar] [CrossRef] [PubMed]
- Levitt, M.; Park, B.H. Water: now you see it, now you don't. Structure 1993, 1, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Morin, C.; Ikeura-Sekiguchi, H.; Tyliszczak, T.; Cornelius, R.; Brash, J.L.; Hitchcock, A.P.; Scholl, A.; Nolting, F.; Appel, G.; Winesett, D.A.; Kaznacheyev, K.; Ade, H. X-ray Spectromicroscopy of immiscible polymer blends: polystyrene-poly(methyl methacrylate). J. Electron Spec. 2001, 121, 203–224. [Google Scholar] [CrossRef]
- Haemostasis and Thrombosis; Bloom, A.L.; Thomas, P.T. (Eds.) Churchill Livingstone: New York, NY, USA, 1987.
- Morin, C.; Hitchcock, A.P.; Cornelius, R.M.; Brash, J.L.; Scholl, A.; Doran, A. Selective adsorption of protein on polymer surfaces studied by soft X-ray photoemission electron microscopy. J. Electron Spectrosc. 2004, 137–140, 785–794. [Google Scholar]
- Peters, T., Jr. All about Albumin: Biochemistry, Genetics, and Medical Applications; Academic Press: New York, NY, USA, 1995. [Google Scholar]
- Li, L.; Hitchcock, A.P.; Robar, N.; Cornelius, R.; Brash, J.L.; Scholl, A.; Doran, A. X-ray microscopy studies of protein adsorption on a phase segregated polystyrene/ polymethylmethacrylate surface. I. concentration and exposure time for albumin adsorption. J. Phys. Chem. B 2006, 110, 16763–16773. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hitchcock, A.P.; Cornelius, R.; Brash, J.L.; Scholl, A.; Doran, A. X-ray microscopy studies of protein adsorption on a phase segregated polystyrene/ polymethylmethacrylate surface, II pH dependence of albumin adsorption. J. Phys. Chem. B 2008, 112, 2150–2158. [Google Scholar] [CrossRef] [PubMed]
- Leung, B.O.; Hitchcock, A.P.; Brash, J.L.; Henklein, P.; Overhage, J.; Hilpert, K.; Hale, J.D.; Hancock, R.E.W. An X-ray spectromicroscopy study of competitive adsorption of protein and peptide onto polystyrene-poly(methyl methacrylate). Biointerphases 2008, 3, F27–F35. [Google Scholar] [CrossRef]
- Leung, B.O.; Hitchcock, A.P.; Brash, J.L.; Scholl, A.; Doran, A. Phase segregation in polystyrene-polylactide blends. Macromolecules 2009, 42, 1679–1684. [Google Scholar] [CrossRef]
- Leung, B.O.; Hitchcock, A.P.; Cornelius, R.; Brash, J.L.; Scholl, A.; Doran, A. An X-ray spectromicroscopy study of protein adsorption to a polystyrene-polylactide blend. Biomacromolecules 2009, 10, 1838–1845. [Google Scholar] [CrossRef] [PubMed]
- Leung, B.O.; Hitchcock, A.P.; Brash, J.L.; Scholl, A.; Doran, A. An X-ray spectromicroscopy study of albumin adsorption to cross-linked polyethylene oxide films. Adv. Biomater. 2010, in press. [Google Scholar]
- Hale, P.; Turgeon, S.; Horny, P.; Lewis, F.; Brack, N.; van Riessen, G.; Pigram, P.; Mantovan, D. X-ray photoelectron emission microscopy and time-of-flight secondary ion mass spectrometry analysis of ultrathin fluoropolymer coatings for stent applications. Langmuir 2008, 24, 7897–7905. [Google Scholar] [CrossRef] [PubMed]
- Hitchcock, A.P.; Morin, C.; Zhang, X.; Araki, T.; Dynes, J.J.; Stöver, H.; Brash, J.L.; Lawrence, J.R.; and Leppard, G.G. Soft X-ray spectromicroscopy of biological and synthetic polymer systems. J. Electron Spectrosc. Relat. Ph. 2005, 144–147, 259–269. [Google Scholar]
- Leung, B.O.; Wang, J.; Brash, J.L.; Hitchcock, A.P. Imaging hydrated albumin on a polystyrene-poly(methyl methacrylate) blend surface with X-ray spectromicroscopy. Langmuir 2009, 25, 13332–13335. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Dupin, D.; Araki, T.; Armes, S.P.; Ade, H. First direct imaging of electrolyte-induced deswelling behavior of pH-responsive microgels in aqueous media using scanning transmission X-ray microscopy. Langmuir 2009, 25, 2588–2592. [Google Scholar] [CrossRef]
- Song, L.; Meng, J.; Zhong, J.; Liu, L.; Dou, X.; Liu, D.; Zhao, X.; Luo, S.; Zhang, Z.; Xiang, Y.; Xu, H.; Zhou, W.; Wu, Z.; Xie, S. Human fibrinogen adsorption onto single-walled carbon nanotube films. Colloid. Surface B 2006, 49, 66–70. [Google Scholar] [CrossRef]
- Andruzzi, L.; Senaratne, W.; Hexemer, A.; Sheets, E.D.; Ilic, B.; Kramer, E.J.; Baird, B.; Ober, C.K. Oligo(ethylene glycol) containing polymer brushes as bioselective surfaces. Langmuir 2005, 21, 2495–2504. [Google Scholar]
- Zubavichus, Y.; Zharnikov, M.; Yang, Y.; Fuchs, O.; Heske, C.; Umbach, E.; Tzvetkov, G.; Netzer, F.P.; Grunze, M. Surface chemistry of ultrathin films of histidine on gold as probed by high-resolution synchrotron photoemission. J. Phys. Chem. B 2005, 109, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Samuel, N.T.; Lee, C.Y.; Gamble, L.J.; Fischer, D.A.; Castner, D.G. NEXAFS characterization of DNA components and molecular-orientation of surface-bound DNA oligomers. J. Electron Spectrosc. Relat. Ph. 2006, 152, 134–142. [Google Scholar] [CrossRef]
- Tobias, W.; Julia, S.; Apte, L.; Gamble, J.; Castner, D.G. Probing the orientation and conformation of α-helix and β-strand model peptides on self-assembled monolayers using sum frequency generation and NEXAFS Spectroscopy. Langmuir 2009. ASAP. [Google Scholar]
- Zubavichus, Y.; Zharnikov, M.; Schaporenko, A.; Grunze, M. NEXAFS study of glycine and glycine-based oligopeptides. J. Electron Spectrosc. Relat. Ph. 2004, 134, 25–33. [Google Scholar] [CrossRef]
- Zubavichus, Y.; Shaporenko, A.; Grunze, M.; Zharnikov, M. NEXAFS spectroscopy of homopolypeptides at all relevant absorption edges: polyisoleucine, polytyrosine, and polyhistidine. J. Phys. Chem. B 2007, 111, 9803–9807. [Google Scholar]
- Zubavichus, Y.; Shaporenko, A.; Grunze, M.; Zharnikov, M. NEXAFS spectroscopy of biological molecules: from amino acids to functional proteins. Nucl. Instrum. Meth. Phys. Res. A 2009, 603, 111–114. [Google Scholar] [CrossRef]
- Zubavichus, Y.; Shaporenko, A.; Grunze, M.; Zharnikov, M. Solid-state near-edge X-ray absorption fine structure spectra of glycine in various charge states. J. Phys. Chem. B 2006, 110, 3420–3427. [Google Scholar] [CrossRef] [PubMed]
- Zubavichus, Y.; Fuchs, O.; Weinhardt, L.; Heske, C.; Umbach, E.; Denlinger, J.D.; Grunze, M. Soft X-ray-induced decomposition of amino acids: an XPS, mass spectrometry, and NEXAFS study. J. Radiat. Res. 2004, 161, 346–358. [Google Scholar] [CrossRef]
- Anders, S.; Padmore, H. A.; Duarte, R. M.; Renner, T.; Stammler, T.; Scholl, A.; Scheinfein, M. R.; Stohr, J.; Seve, L.; Sinkovic, B. Photoemission electron microscope for the study of magnetic materials. Rev. Sci. Instrum. 1999, 70, 3973–3981. [Google Scholar] [CrossRef]
- Wang, J.; Li, L.; Morin, C.; Hitchcock, A.P.; Doran, A.; Scholl, A. Radiation damage in soft X-ray microscopy. J. Electron Spectrosc. Relat. Phenom. 2009, 170, 25–36. [Google Scholar] [CrossRef]
- Hitchcock, A.P.; Stöver, H.D.H.; Croll, L.M.; Childs, R.F. Chemical mapping of polymer microstructure using soft X-ray spectromicroscopy. Australian J. Chem. 2005, 58, 423–432. [Google Scholar]
- Kilcoyne, A.L.D.; Tylisczak, T.; Steele, W.F.; Fakra, S.; Hitchcock, P.; Franck, K.; Anderson, E.; Harteneck, B.; Rightor, E.G.; Mitchell, G.E.; Hitchcock, A.P.; Yang, L.; Warwick, T.; Ade, H. Interferometer-controlled scanning transmission X-ray microscopes at the Advanced Light Source. J. Synchrotron Radiat. 2003, 10, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Warwick, T.; Ade, H.; Kilcoyne, A.L.D.; Kritscher, M.; Tylisczcak, T.; Fakra, S.; Hitchcock, A.P.; Hitchcock, P.; Padmore, H.A. A new bend-magnet beamline for scanning transmission X-ray microscopy at the Advanced Light Source. J. Synchrotron Radiat. 2002, 9, 254–257. [Google Scholar] [CrossRef] [PubMed]
- aXis2000 is free for noncommercial use. It is written in Interactive Data Language (IDL) and available online: http://unicorn.mcmaster.ca/aXis2000.html (accessed on 5 June 2010).
- Jacobsen , C.; Wirick, S.; Flynn, G. Soft X-ray Spectroscopy from image sequences with Sub-100 nm spatial resolution. J. Microscopy 2000, 197, 173. [Google Scholar]
- Strang, G. Linear Algebra and Its Applications; Harcourt Bracourt, Jovanovich: San Diego, CA, USA, 1988. [Google Scholar]
- Koprinarov, I.N.; Hitchcock, A.P.; McCrory, C.T.; Childs, R.F. Quantitative mapping of structured polymeric systems using singular value decomposition analysis of soft X-ray images. J. Phys. Chem. B 2002, 106, 5358–5364. [Google Scholar] [CrossRef]
- Campbell, I.M. Introduction to Synthetic Polymers; Oxford University Press: New York, NY, USA, 1994. [Google Scholar]
- Wu, S. Surface and interfacial tensions of polymer melts. II. Poly(methyl methacrylate), poly(n-butyl methacrylate), and polystyrene. J. Phys. Chem. 1970, 74, 632–638. [Google Scholar] [CrossRef]
- Stewart-Ornstein, J.; Hitchcock, A.P.; Hernandez-Cruz, D.; Henklein, P.; Overhage, J.; Hilpert, K.; Hale, J.D.; Hancock, R.E.W. Using intrinsic X-ray absorption spectral differences to identify and map peptides and proteins. J. Phys. Chem. B 2007, 111, 7691. [Google Scholar] [CrossRef] [PubMed]
- Gros, G.; Forster, R.E.; Lin, L. The carbamate reaction of glycylglycine, plasma, and tissue extracts evaluated by a pH stopped flow apparatus. J. Biol. Chem. 1976, 251, 4398–4407. [Google Scholar] [PubMed]
- Liu, Y.; Messmer, M.C. Surface structures and segregation of polystyrene/poly(methyl methacrylate) blends studied by sum-frequency (SF) spectroscopy. J. Phys. Chem. B 2003, 107, 9774–9779. [Google Scholar] [CrossRef]
- Foster, J.F. Albumin Structure, Function and Uses; Rosenoer, V.M., Oratz, M., Rothschild, M.A., Eds.; Pergamon: Oxford, UK, 1977; pp. 53–84. [Google Scholar]
- Fogh-Andersen, N.; Bjerrum, P.J.; Siggaard-Andersen, O. Ionic binding, net charge, and Donnan effect of human serum albumin as a function of pH. Clin. Chem. 1993, 39, 48–52. [Google Scholar] [PubMed]
- Zubavichus, Y.; Shaporenko, A.; Grunze, M.; Zharnikov, M. Innershell absorption spectroscopy of amino acids at all relevant absorption edges. J. Phys. Chem C. 2005, 109, 6998. [Google Scholar] [CrossRef]
- Hilpert, K.; Volkmer-Engert, R.; Walter, T.; Hancock, R.E.W. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 2005, 23, 1008. [Google Scholar] [CrossRef] [PubMed]
- Ragauskas, A.J.; Williams, C.K.; Davison, B.H.; Britovsek, G.; Cairney, J.; Eckert, C.A.; Frederick, W.J.; Hallet, J.P.; Leak, D.J.; Liotta, C.L.; Mielenz, J.R.; Murphy, R.; Templer, R.; Tschaplinksi, T. The path forward for biofuels and biomaterials. Science 2006, 311, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Athanasiou, K.A.; Niederauer, G.G.; Agrawal, C.M. Sterilization, toxicity, biocompatibility and clinical applications of polylactic acid/ polyglycolic acid copolymers. Biomaterials 1996, 17, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Hutmacher, D.W. Scaffolds in tissue engineering bone and cartilage. Biomaterials 2000, 21, 2529–2543. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Teply, B.A.; Jeong, S.Y.; Yim, C.H.; Ho, D.; Sherifi, I.; Jon, S.; Farokhzad, O.C.; Khademhosseini, A.; Langer, R.S. Magnetically responsive polymeric microparticles for oral delivery of protein drugs. Pharmaceut. Res. 2006, 23, 557–564. [Google Scholar] [CrossRef]
- Mohamad, A.; Gordon, S.H.; Biresaw, G.J. Poly(lactic acid)/polystyrene bioblends characterized by thermogravimetric analysis, differential scanning calorimetry, and photoacoustic infrared spectroscopy. J. Appl. Polym. Sci. 2007, 106, 1689–1696. [Google Scholar] [CrossRef]
- Steinmann, S.; Gronski, W.; Friedrich, C. Cocontinuous polymer blends: influence of viscosity and elasticity ratios of the constituent polymers on phase inversion. Polymer 2001, 42, 6619–6629. [Google Scholar] [CrossRef]
- Utracki, L.A. On the viscosity-concentration dependence of immiscible polymer blends. Rheol. 1991, 35, 1615–1637. [Google Scholar] [CrossRef]
- Su, T.J.; Lu, J.R.; Thomas, R.K.; Cui, Z.F.; Penfold, J.J. The adsorption of lysozyme at the silica–water interface: a neutron reflection study. Colloids Interface Sci. 1998, 203, 419–429. [Google Scholar] [CrossRef]
- Pache, S.; Voros, J.; Griesser, J.H.; Spencer, N.D.; Textor, M. Effects of ionic strength and surface charge on protein adsorption at PEGylated surfaces. J. Phys. Chem B 2008, 109, 17545–17552. [Google Scholar] [CrossRef]
- Ramsden, J.J.; Prenosil, J.E. Effect of ionic strength on protein adsorption kinetics. J. Phys. Chem 1994, 98, 5376–5381. [Google Scholar] [CrossRef]
- Majeti, N.V.; Ravi, K. A review of chitin and chitosan applications. React. Funct. Polym. 2000, 46, 1–27. [Google Scholar]
- Solomon, D.; Lehmann, J.; Kinyangi, J.; Liang, B.; Heymann, K.; Dathe, L.; Hanley, K.; Wirick, S.; Jacobsen, C. Carbon (1s) NEXAFS spectroscopy of biogeochemically relevant reference organic compounds. SSSAJ 2009, 73, 1817–1830. [Google Scholar] [CrossRef]
- Merrill, E.W.; Dennison, K.A.; Sung, C. Partitioning and diffusion of solutes in hydrogels of poly(ethylene oxide). Biomaterials 1993, 14, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Quinn, C.P.; Patha, C.P.; Heller, A.; Hubbell, J.A. Photo-crosslinked copolymers of 2-hydroxyethyl methacrylate, poly(ethylene glycol) tetra-acrylate and ethylene dimethacrylate for improving biocompatibility of biosensors. Biomaterials 1995, 16, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Stringer, J.L.; Peppas, N.A. Diffusion of small molecular weight drugs in radiation-crosslinked poly(ethylene oxide) hydrogels. J. Control. Release 1996, 42, 195–202. [Google Scholar] [CrossRef]
- Doycheva, M.; Stamenova, R.; Zvetkov, V.; Tsvetanov, C.B. UV irradiation-induced crosslinking of solid poly(ethylene oxide) modified with tetraalkyl ammonium salt. Polymer 1998, 39, 6715–6721. [Google Scholar] [CrossRef]
- Mellott, M.B.; Searcy, K.; Pishko, M.V. Release of protein from highly cross-linked hydrogels of poly(ethylene glycol) diacrylate fabricated by UV polymerization. Biomaterials 2001, 22, 929–941. [Google Scholar] [CrossRef] [PubMed]
- Kofinas, P.; Athanassiou, V.; Merrill, E.W. Hydrogels prepared by electron irradiation of poly(ethylene oxide) in water solution: unexpected dependence of cross-link density and protein diffusion coefficients on initial PEO molecular weight. Biomaterials 1996, 17, 1547–1550. [Google Scholar]
- Doycheva, M.; Petrova, E.; Stamenova, R.; Tsvetanov, C.; Riess, G. Ultraviolet-induced crosslinking of solid poly(ethylene oxide). Macromol. Mater. Eng. 2004, 289, 676–680. [Google Scholar] [CrossRef]
- Doytcheva, M.; Dotcheva, D.; Stamenova, R.; Tsvetanov, C. UV-Induced cross-linking of poly(ethylene oxide) in Aaueous solution. Macrmol. Mater. Eng. 2001, 286, 30–33. [Google Scholar] [CrossRef]
- Dimitrov, M.; Dotcheva, D.; Lambov, N. Preparation and characterization of polyethilene oxide hydrogels with cytisine. Acta Pharm. Turcica 2004, 46, 49–54. [Google Scholar]
- Petrov, P.; Bozukov, M.; Burkhardt, M.; Muthukrishnan, S.; Müller, A.H.E.; Tsvetanov, C.B. Stabilization of polymeric micelles with a mixed poly(ethylene oxide)/poly(2-hydroxyethyl methacrylate) shell by formation of poly(pentaerythritol tetraacrylate) nanonetworks within the micelles. J. Mater. Chem. 2006, 16, 2192–2199. [Google Scholar] [CrossRef]
- Lensen, M. C.; Mela, P.; Mourran, A.; Groll, J.; Heuts, J.; Rong, H.; Möller, M. Micro- and nanopatterned star poly(ethylene glycol) (PEG) materials prepared by UV-based imprint lithography. Langmuir 2007, 23, 7841–7846. [Google Scholar] [CrossRef] [PubMed]
- Hale, P.; Turgeon, S.; Horny, P.; Lewis, F.; Brack, N.; van Riessen, G.; Pigram, P.; Mantovan, D. X-ray photoelectron emission microscopy and time-of-flight secondary ion mass spectrometry analysis of ultrathin fluoropolymer coatings for stent applications. Langmuir 2008, 24, 7897–7905. [Google Scholar] [CrossRef] [PubMed]
- Lewis, F.; Horny, P.; Hale, P.; Turgeon, S.; Tatoulian, M.; Mantovani, D. Study of the adhesion of thin plasma fluorocarbon coatings. J. Phys. D: Appl. Phys. 2008, 41, 045310–045317. [Google Scholar] [CrossRef]
- Hitchcock, A.P.; Morin, C.; Heng, Y.M.; Cornelius, R.M.; Brash, J.L. Towards practical soft X-ray spectromicroscopy of biomaterials. J. Biomater. Sci. Polymer Ed. 2002, 13, 919–938. [Google Scholar] [CrossRef]
- Hitchcock, A.P.; Koprinarov, I.; Tyliszczak, T.; Rightor, E.G.; Mitchell, G.E.; Dineen, M.T.; Hayes, F.; Lidy, W.; Priester, R.D.; Urquhart, S.G.; Smith, A.P.; Ade, H. Copolymer polyol particles in polyurethanes studied by soft X-ray spectromicroscopy. Ultramicroscopy 2001, 88, 33–49. [Google Scholar]
- Hook, F.; Voros, J.; Rodahl, M.; Kurrat, R.; Boni, P.; Ramsden, J.J.; Textor, M.; Spencer, N.D.; Tengvall, P.; Gold, J.; Kasemo, B. A comparative study of protein adsorption on titanium oxide surfaces using in situ ellipsometry, optical waveguide lightmode spectroscopy, and quartz crystal microbalance/dissipation. Colloids Surf. B 2002, 24, 155–170. [Google Scholar] [CrossRef]
- Rechendorff, K.; Hovgaard, M.B.; Foss, M.; Zhdanov, V.P.; Besenbacher, F. Enhancement of protein adsorption induced by surface roughness. Langmuir 2006, 22, 10885–10888. [Google Scholar] [CrossRef] [PubMed]
- Sugio, S.; Kashima, A.; Mochizuki, S.; Noda, M.; Kobayashi, K. Crystal structure of human serum albumin at 2.5 A resolution. Protein Eng. 1999, 12, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Su, T.J.; Lu, J.R.; Thomas, R.K.; Cui, Z.F.; Penfold, J. The conformational structure of bovine serum albumin layers adsorbed at the silica−water Interface. J. Phys. Chem. B 1998, 102, 8100–8108. [Google Scholar] [CrossRef]
- Morris, G.E.; Vincent, B.; Snowden, M.J.J. Adsorption of lead ions onto N-Isopropylacrylamide and acrylic acid copolymer microgels. Colloid Interface Sci. 1997, 190, 198–205. [Google Scholar] [CrossRef]
- Snowden, M.J.; Thomas, D.; Vincent, B. Use of colloidal microgels for the absorption of heavy metal and other ions from aqueous solution. Analyst 1993, 118, 1367–1369. [Google Scholar] [CrossRef]
- Holtz, J.H.; Asher, S.A. Polymerized colloidal crystal hydrogel films as intelligent chemical sensing materials. Nature 1997, 389, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Jeong, B.; Bae, Y.H.; Lee, D.S.; Kim, S.W. Biodegradable block copolymers as injectable drug-delivery systems. Nature 1997, 388, 860–862. [Google Scholar] [CrossRef] [PubMed]
- Murthy, N.; Thng, Y.X.; Schuck, S.; Xu, M.C.; Frechet, J.M.J. A novel strategy for encapsulation and release of proteins: hydrogels and microgels with acid-labile acetal cross-linkers. J. Am. Chem. Soc. 2002, 124, 12398–12399. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.; Lee, H.; Chmielewski, J.; Lyon, L.A. Folate-mediated cell targeting and cytotoxicity using rhermoresponsive microgels. J. Am. Chem. Soc. 2004, 126, 10258–10259. [Google Scholar] [CrossRef] [PubMed]
- Al-Azzam, W.; Pastrana, E.A.; Ferrer, Y.; Huang, Q.; Schweitzer-Stenner, R.; Griebenow, K. Structure of poly(ethylene glycol)-modified horseradish peroxidase in organic solvents: infrared amide I spectral changes upon protein dehydration are largely caused by protein structural changes and not by water removal per se. Biophys. J. 2002, 83, 3637–3651. [Google Scholar] [CrossRef] [PubMed]
- Griebenow, K.; Klibanov, A.M. Lyophilization-induced reversible changes in the secondary structure of proteins. Proc. Natl. Acad. Sci. USA 1995, 92, 10969. [Google Scholar] [CrossRef] [PubMed]
© 2010 by the authors. This article is an Open Access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Leung, B.O.; Brash, J.L.; Hitchcock, A.P. Characterization of Biomaterials by Soft X-Ray Spectromicroscopy. Materials 2010, 3, 3911-3938. https://doi.org/10.3390/ma3073911
Leung BO, Brash JL, Hitchcock AP. Characterization of Biomaterials by Soft X-Ray Spectromicroscopy. Materials. 2010; 3(7):3911-3938. https://doi.org/10.3390/ma3073911
Chicago/Turabian StyleLeung, Bonnie O., John L. Brash, and Adam P. Hitchcock. 2010. "Characterization of Biomaterials by Soft X-Ray Spectromicroscopy" Materials 3, no. 7: 3911-3938. https://doi.org/10.3390/ma3073911



