Single Voxel Proton Spectroscopy for Neurofeedback at 7 Tesla
Abstract
:1. Introduction
2. Methods Section
2.1. Spectral Processing of the Water Signal
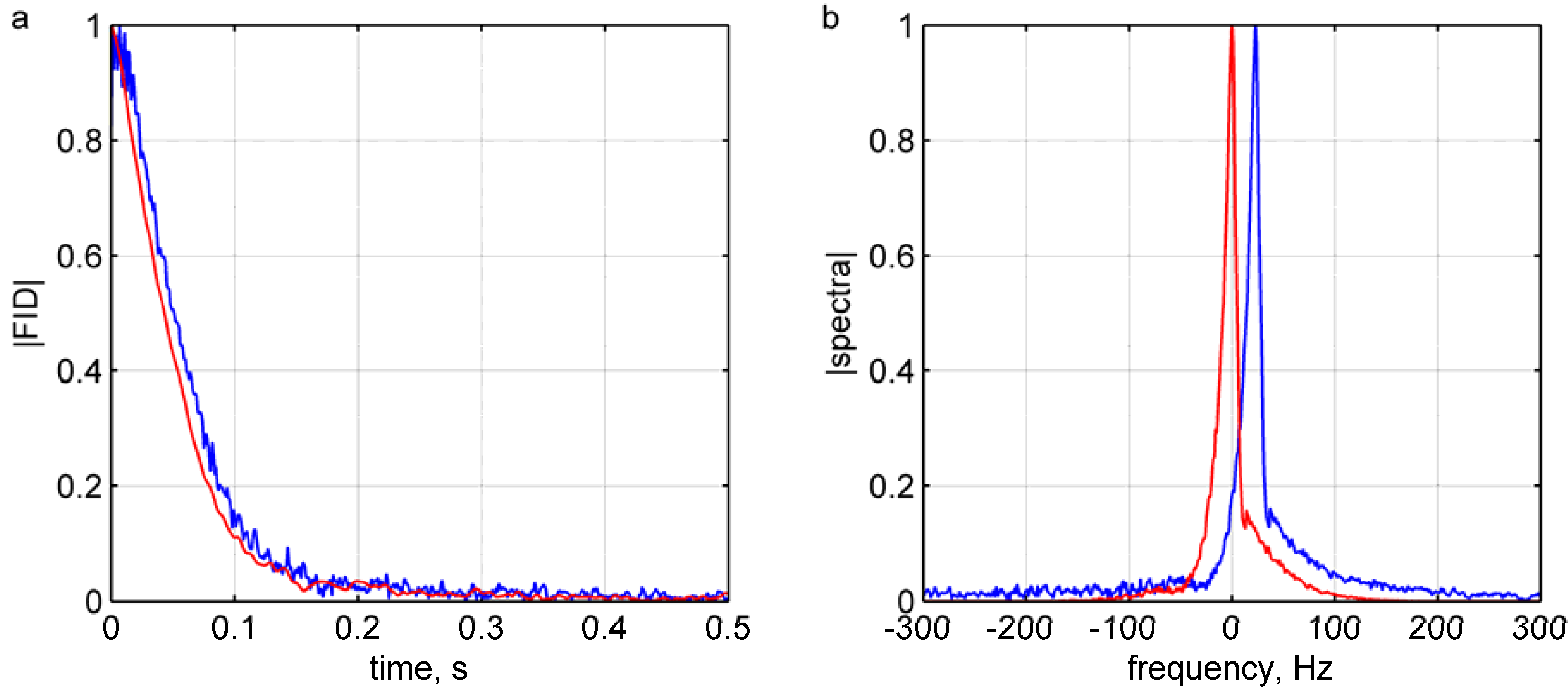

2.2. Real-Time Feedback Signal Processing
2.2.1. EMA as a High-Pass Filter
2.2.2. Non-Linear Modified Kalman Filter as Low-Pass Filter and Spike Detector
2.2.3. Normalization of the Feedback Signal
2.3. Method Performance Assessment Parameters
3. Experimental Section
3.1. Neurofeedback Data Acquisition and Preprocessing
3.2. Experimental Design
4. Results and Discussion
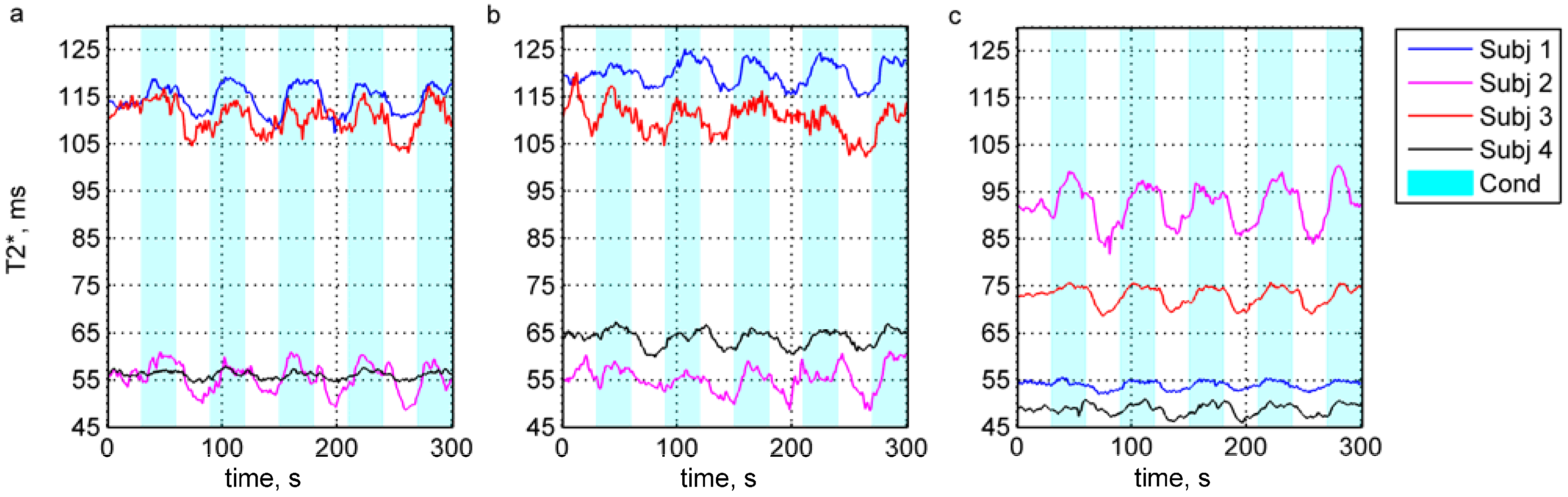
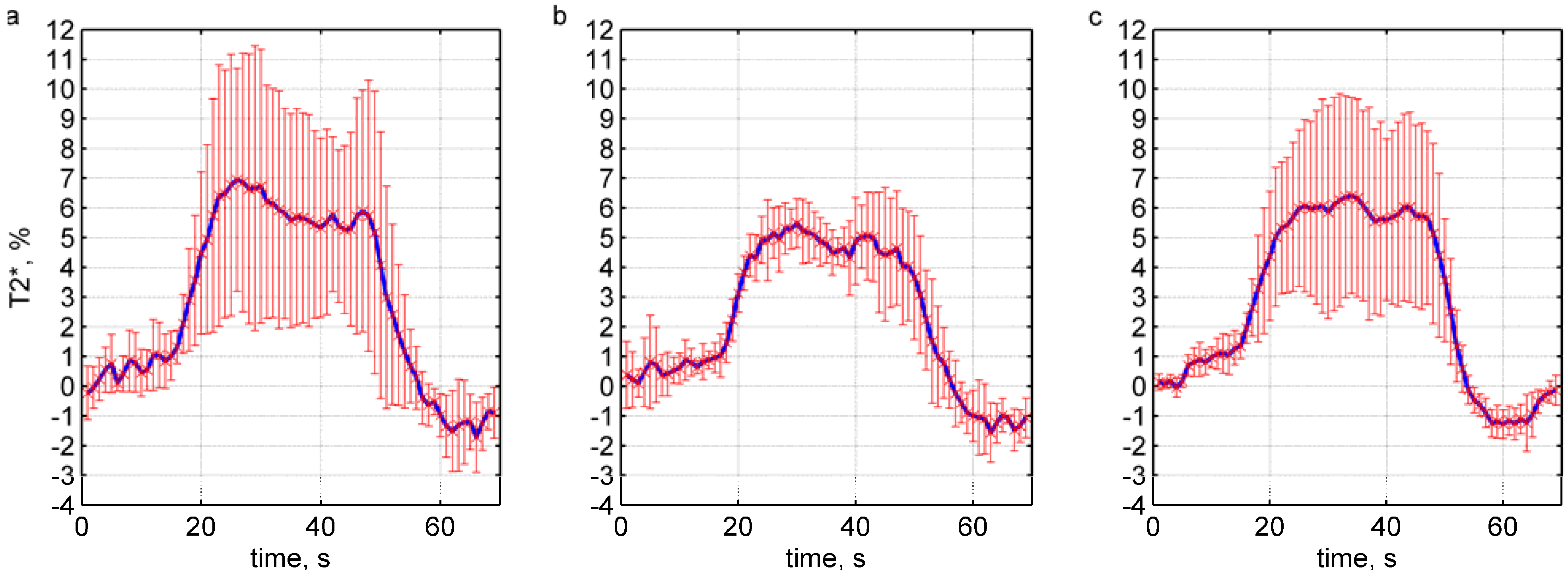
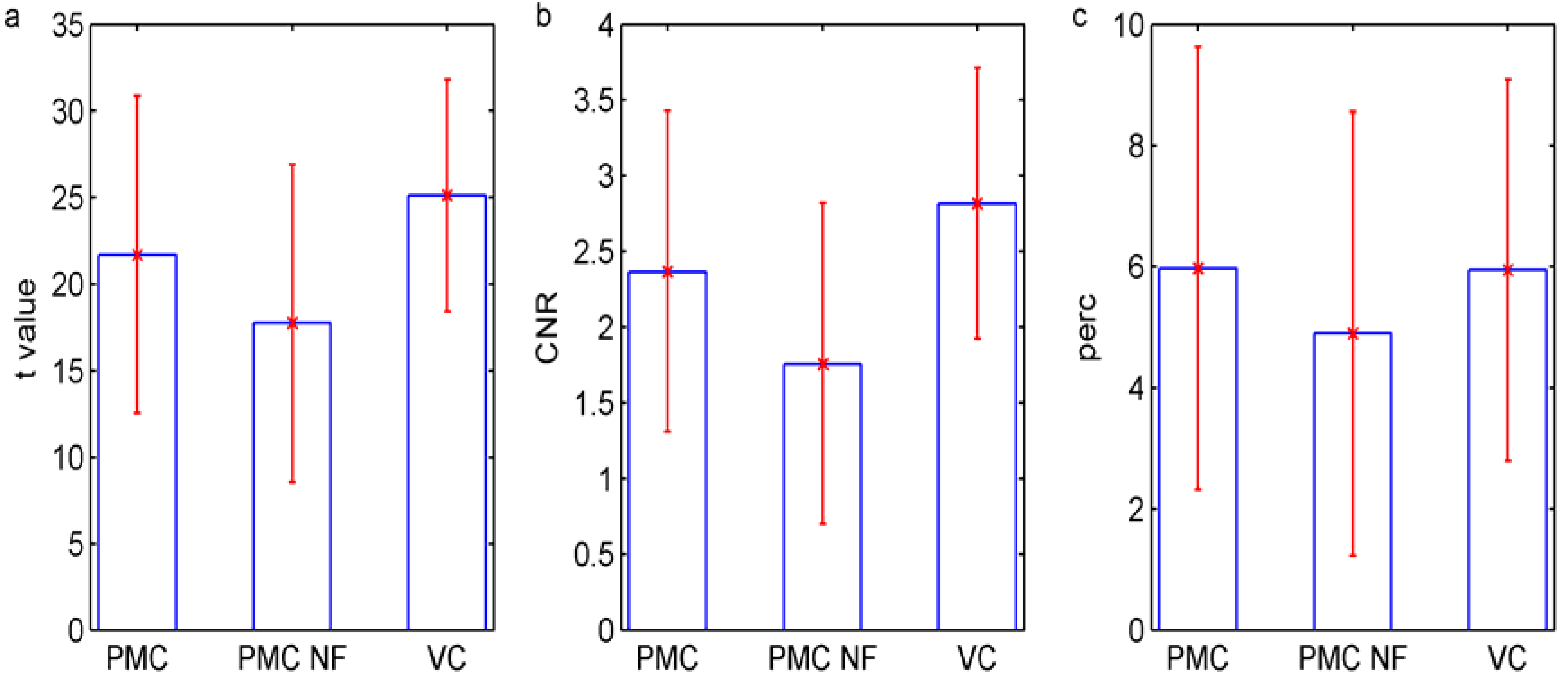

5. Conclusions
Supplementary Materials
Acknowledgments
References
- Goebel, R. Cortex-based real-time fMRI. Neuroimage 2001, 13, S129. [Google Scholar] [CrossRef]
- Weiskopf, N.; Mathiak, K.; Bock, S.W.; Scharnowski, F.; Veit, R.; Grodd, W.; Goebel, R.; Birbaumer, N. Principles of a Brain-Computer Interface (BCI) based on real-time functional magnetic resonance imaging (fMRI). IEEE Trans. Biomed. Eng. 2004, 51, 966–970. [Google Scholar] [CrossRef] [PubMed]
- Weiskopf, N.; Veit, R.; Erb, M.; Mathiak, K.; Grodd, W.; Goebel, R.; Birbaumer, N. Physiological self-regulation of regional brain activity using real-time functional magnetic resonance imaging (fMRI): Methodology and exemplary data. Neuroimage 2003, 19, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Weiskopf, N.; Sitaram, R.; Josephs, O.; Veit, R.; Scharnowski, F.; Goebel, R.; Birbaumer, N.; Deichmann, R.; Mathiak, K. Real-time functional magnetic resonance imaging: methods and applications. Magn. Reson. Imaging 2007, 25, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- deCharms, R.C.; Maeda, F.; Glover, G.H.; Ludlow, D.; Pauly, J.M.; Soneji, D.; Gabrieli, J.D.E.; Mackey, S.C. Control over brain activation and pain learned by using real-time functional MRI. PNAS 2005, 102, 18626–18631. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.; Shimojo, J.P.; O’Doherty, J.P. Direct instrumental conditioning of neural activity using functional magnetic resonance imaging-derived reward feedback. J. Neurosci. 2007, 27, 7498–7507. [Google Scholar] [CrossRef] [PubMed]
- Caria, A.; Veit, R.; Sitaram, R.; Lotze, M.; Weiskopf, N.; Grodd, W.; Birbaumer, N. Regulation of anterior insular cortex activity using real-time fMRI. Neuroimage 2007, 35, 1238–1246. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.S.; Lee, J.H.; O’Leary, H.; Panych, L.P.; Jolesz, F.A. Neurofeedback fMRI-mediated learning and consolidation of regional brain activation during motor imagery. Int. J. Imaging Syst. Technol. 2008, 18, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Rota, G.; Sitaram, R.; Veit, R.; Erb, M.; Weiskopf, N.; Dogil, G.; Birbaumer, N. Self-regulation of regional cortical activity using real-time fMRI: the right inferior frontal gyrus and linguistic processing. Hum. Brain Mapp. 2009, 30, 1605–1614. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Ryu, J.; Jolesz, F.A.; Cho, Z.H.; Yoo, S.S. Brain-machine interface via real-time fMRI: Preliminary study on thought-controlled robotic arm. Neurosci. Lett. 2009, 450, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Birbaumer, N.; Murguialday, A.R.; Weber, C.; Montoya, P. Neurofeedback and brain-computer interface clinical applications. Int. Rev. Neurobiol. 2009, 86, 107–117. [Google Scholar] [PubMed]
- Johnston, S.J.; Boehm, S.G.; Healy, D.; Goebel, R.; Linden, D.E.J. Neurofeedback: A promising tool for the self-regulation of emotion networks. Neuroimage 2010, 49, 1066–1072. [Google Scholar] [CrossRef] [PubMed]
- LaConte, S.M. Decoding fMRI brain states in real-time. Neuroimage 2011, 56, 440–454. [Google Scholar] [CrossRef] [PubMed]
- Lubar, J.E.; Swartwood, M.O.; Swartwood, J.N.; O’Donnell, P.H. Evaluation of the effectiveness of EEG neurofeedback training for ADHD in a Clinical Setting as measured by changes in TOVA scores, behavioral ratings and WISC-R performance. Biofeedback Self-Regul. 1995, 20, 83–99. [Google Scholar] [CrossRef] [PubMed]
- Kotchoubey, B.; Strehl, U.; Uhlmann, C.; Holzapfel, S.; König, M.; Fröscher, W.; Blankenhorn, V.; Birbaumer, N. Modulation of slow cortical potentials in patients with intractable epilepsy. Epilepsia 2001, 42, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Villringer, A.; Chance, B. Noninvasive optical spectroscopy and imaging of human brain function. Trends Neurosci. 1997, 20, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Rolfe, P. In vivo near-infrared spectroscopy. Annu. Rev. Biomed. Eng. 2000, 2, 715–754. [Google Scholar] [CrossRef] [PubMed]
- Abdelnour, A.F.; Huppert, T. Real-time imaging of human brain function by near-infrared spectroscopy using an adaptive general linear model. Neuroimage 2009, 46, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Bray, S.; Reiss, A.L. Functional near infrared spectroscopy (NIRS) signal improvement based on negative correlation between oxygenated and deoxygenated hemoglobin dynamics. Neuroimage 2009, 49, 3039–3046. [Google Scholar] [CrossRef] [PubMed]
- Hennig, J.; Ernst, T.; Speck, O. Detection of brain activation using oxygenation sensitive functional spectroscopy. Magn. Reson. Med. 1994, 31, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Hennig, J.; Janz, C.; Speck, O.; Ernst, T. Functional spectroscopy of brain activation following a single light pulse: Examinations of the mechanism of the fast initial response. Int. J. Imaging Syst. Technol. 1995, 6, 203–208. [Google Scholar] [CrossRef]
- Richards, T.L.; Dager, S.R.; Posse, S. Functional MR spectroscopy of the brain. Neuroimaging Clin. N. Am. 1998, 8, 823–824. [Google Scholar] [PubMed]
- Mulkern, R.V.; Chen, N.; Oshio, K.; Panych, L.P.; Rybicki, F.J.; Gambarota, G. Fast spectroscopic imaging strategies for potential applications in fMRI. Magn. Reson. Imaging 2004, 22, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Ebel, A.; Ji, J.X.; Schuff, N. Spectral phase-corrected GRAPPA reconstruction of three-dimensional echo-planar spectroscopic imaging (3D-EPSI). Magn. Reson. Med. 2007, 57, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.H.; Tsai, S.Y.; Otazo, R.; Caprihan, A.; Wald, L.L.; Belliveau, J.W.; Posse, S. Sensitivity-Encoded (SENSE) Proton Echo-Planar Spectroscopic Imaging (PEPSI) in the human brain. Magn. Res. Med. 2007, 57, 249–257. [Google Scholar] [CrossRef]
- Chen, N.K.; Oshio, K.; Panych, L.P.; Rybicki, F.J.; Mulkern, R.V. Spatially selective T2 and T2* measurement with line-scan echo-planar spectroscopic imaging. J. Magn. Reson. 2004, 171, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Golay, X.; Gillen, J.; van Zijl, P.C.; Barker, P.B. Scan time reduction in proton magnetic resonance spectroscopic imaging of the human brain. Magn. Reson. Med. 2002, 47, 384–387. [Google Scholar] [CrossRef] [PubMed]
- Dydak, U.; Weiger, M.; Pruessmann, K.P.; Meier, D.; Boesiger, P. Sensitivity-encoded spectroscopic imaging. Magn. Reson. Med. 2001, 46, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Posse, S.; Tedeschi, G.; Risinger, R.; Ogg, R.; Le Bihan, D. High speed 1H spectroscopic imaging in human brain by echo planar spatial-spectral encoding. Magn. Reson. Med. 1995, 33, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Bonekamp, D.; Smith, M.A.; Zhua, H.; Barker, P.B. Quantitative SENSE-MRSI of the human brain. Magn. Reson. Imaging 2010, 28, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.L.; Schlemmer, H.; Yeung, D.K.W.; Wilhelm, T.; Bachert, P. Comparison of magnetic resonance spectroscopic imaging and single voxel magnetic resonance spectroscopy for suspected recurrent brain tumour or radiation necrosis. J. HK Coll. Radiol. 2001, 4, 259–263. [Google Scholar]
- Mekle, R.; Mlynarik, V.; Gambarota, G.; Hergt, M.; Krueger, G.; Gruetter, R. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn. Reson. Med. 2009, 61, 1279–1285. [Google Scholar] [CrossRef] [PubMed]
- Dydak, U.; Schär, M. MR spectroscopy and spectroscopic imaging: Comparing 3.0 T versus 1.5 T. Neuroimag Clin. N. Am. 2006, 16, 269–283. [Google Scholar] [CrossRef]
- Costanzo, A.; Trojsi, F.; Tosetti, M.; Giannatempo, G.M.; Nemore, F.; Piccirillo, M.; Sonavita, S.; Tedeschi, G.; Scarabino, T. High-field proton MRS of human brain. Euro. J. Radiol. 2003, 48, 146–153. [Google Scholar] [CrossRef]
- Mangia, S.; Tkac, I.; Gruetter, R.; Van De Moortele, P.F.; Giove, F.; Maraviglia, B.; Kamil Ugurbil, K. Sensitivity of single-voxel 1H-MRS in investigating the metabolism of the activated human visual cortex at 7 T. Magn. Reson. Imaging 2006, 24, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Diedrichsen, J.; Shadmehr, R. Detecting and adjusting for artifacts in fMRI time series data. Neuroimage 2005, 27, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Hinds, O.; Ghosh, S.; Thompson, T.W.; Yoo, J.J.; Gabrieli, S.W.; Triantafyllou, C.; Gabrieli, J.D.E. Computing moment-to-moment BOLD activation for real-time neurofeedback. Neuroimage 2010, 54, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Ultra High Field Magnetic Resonance Imaging; Robitaille, P.-M.; Berliner, L. (Eds.) Biological Magnetic Resonance; Vol. 26, Springer: New York, NY, USA, 2006.
- Poser, B.A.; Norris, D.G. Investigating of benefits of multi-echo EPI for fMRI at 7T. Neuroimage 2009, 45, 1162–1172. [Google Scholar] [CrossRef] [PubMed]
- Simmons, A.; Moore, E.; Williams, S.C. Quality control for functional magnetic resonance imaging using automated data analysis and Shewhart charting. Magn. Reson. Med. 1999, 41, 1274–1278. [Google Scholar] [CrossRef] [PubMed]
- Stöcker, T.; Schneider, F.; Klein, M.; Habel, U.; Kellermann, T.; Zilles, K.; Shah, N.J. Automated quality assurance routines for fMRI data applied to a multicenter study. Hum. Brain Mapp. 2005, 237–246. [Google Scholar]
- Koush, Y.; Zvyagintsev, M.; Dyck, M.; Mathiak, K.A.; Mathiak, K. Signal quality and Bayesian signal processing in neurofeedback based on real-time fMRI. Neuroimage 2011. Epub ahead of print. [Google Scholar] [CrossRef]
- Roberts, S.W. Control chart tests based on geometric moving averages. Technometrics 1959, 1, 239–250. [Google Scholar] [CrossRef]
- Uludag, K.; Dubowitz, D.J.; Buxton, R.B. Basic Principles of Functional MRI. In Clinical MRI; Edelman, R., Hesselink, J., Zlatkin, M., Eds.; Elsevier: San Diego, CA, USA, 2005; pp. 249–287. [Google Scholar]
- Robinson, P.A.; Drysdale, P.M.; Van der Merwe, H.; Kyriakou, E.; Rigozzi, M.K.; Germanoska, B.; Renniea, C.J. BOLD responses to stimuli: Dependence on frequency, stimulus form, amplitude, and repetition rate. Neuroimage 2006, 31, 585–599. [Google Scholar] [CrossRef] [PubMed]
- Kannurpatti, S.S.; Biswal, B.B.; Kim, Y.R.; Rosen, B.R. Spatio-temporal characteristics of low-frequency BOLD signal fluctuations in isoflurane-anesthetized rat brain. Neuroimage 2008, 40, 1738–1747. [Google Scholar] [CrossRef] [PubMed]
- Kalman, R.E. A new approach to linear filtering and prediction problems. Trans. ASME J. Basic Eng. 1960, 82, 35–45. [Google Scholar] [CrossRef]
- Vaseghi, S.V. Advanced Digital Signal Processing and Noise Reduction, 3rd ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2006; pp. 188–196. [Google Scholar]
- Grewal, M.S.; Andrews, A.P. Kalman Filtering: Theory and practice using Matlab, 2nd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2008. [Google Scholar]
- Weiskopf, N.; Scharnowski, F.; Veit, R.; Goebel, R.; Birbaumer, N.; Mathiak, K. Self-regulation of local brain activity using real-time functional magnetic resonance imaging (fMRI). J. Physiol. 2004, 98, 357–373. [Google Scholar]
- Duong, T.Q.; Yacoub, E.; Adriany, G.; Hu, X.; Ugurbil, K.; Kim, S.G. Microvascular BOLD contribution at 4 and 7T in the human brain: gradient-echo and spin-echo fMRI with suppression of blood effects. Magn. Reson. Med. 2003, 49, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Yacoub, E.; Schmuel, A.; Logothetis, N.; Ugurbil, K. Robust detection of ocular dominance columns in humans using Hahn Spin Echo BOLD functional MRI at 7 Tesla. Neuroimage 2007, 4, 1161–1177. [Google Scholar] [CrossRef]
- Hess, A.T.; Tisdal, M.D.; Andronesi, O.C.; Meintjes, E.M.; van der Kouwe, A.J.W. Real-time Motion and B0 corrected single voxel spectroscopy using volumetric navigators. Magn. Reson. Med. 2011, 66, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Zaitsev, M.; Speck, O.; Hennig, J.; Büchert, M. Single-voxel MRS with prospective motion correction and retrospective frequency correction. NMR Biomed. 2010, 23, 325–332. [Google Scholar] [PubMed]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Koush, Y.; Elliott, M.A.; Mathiak, K. Single Voxel Proton Spectroscopy for Neurofeedback at 7 Tesla. Materials 2011, 4, 1548-1563. https://doi.org/10.3390/ma4091548
Koush Y, Elliott MA, Mathiak K. Single Voxel Proton Spectroscopy for Neurofeedback at 7 Tesla. Materials. 2011; 4(9):1548-1563. https://doi.org/10.3390/ma4091548
Chicago/Turabian StyleKoush, Yury, Mark A. Elliott, and Klaus Mathiak. 2011. "Single Voxel Proton Spectroscopy for Neurofeedback at 7 Tesla" Materials 4, no. 9: 1548-1563. https://doi.org/10.3390/ma4091548



