Pressure Dependence of 15N Chemical Shifts in Model Peptides Ac-Gly-Gly-X-Ala-NH2
Abstract
:1. Introduction
2. Results and Discussion
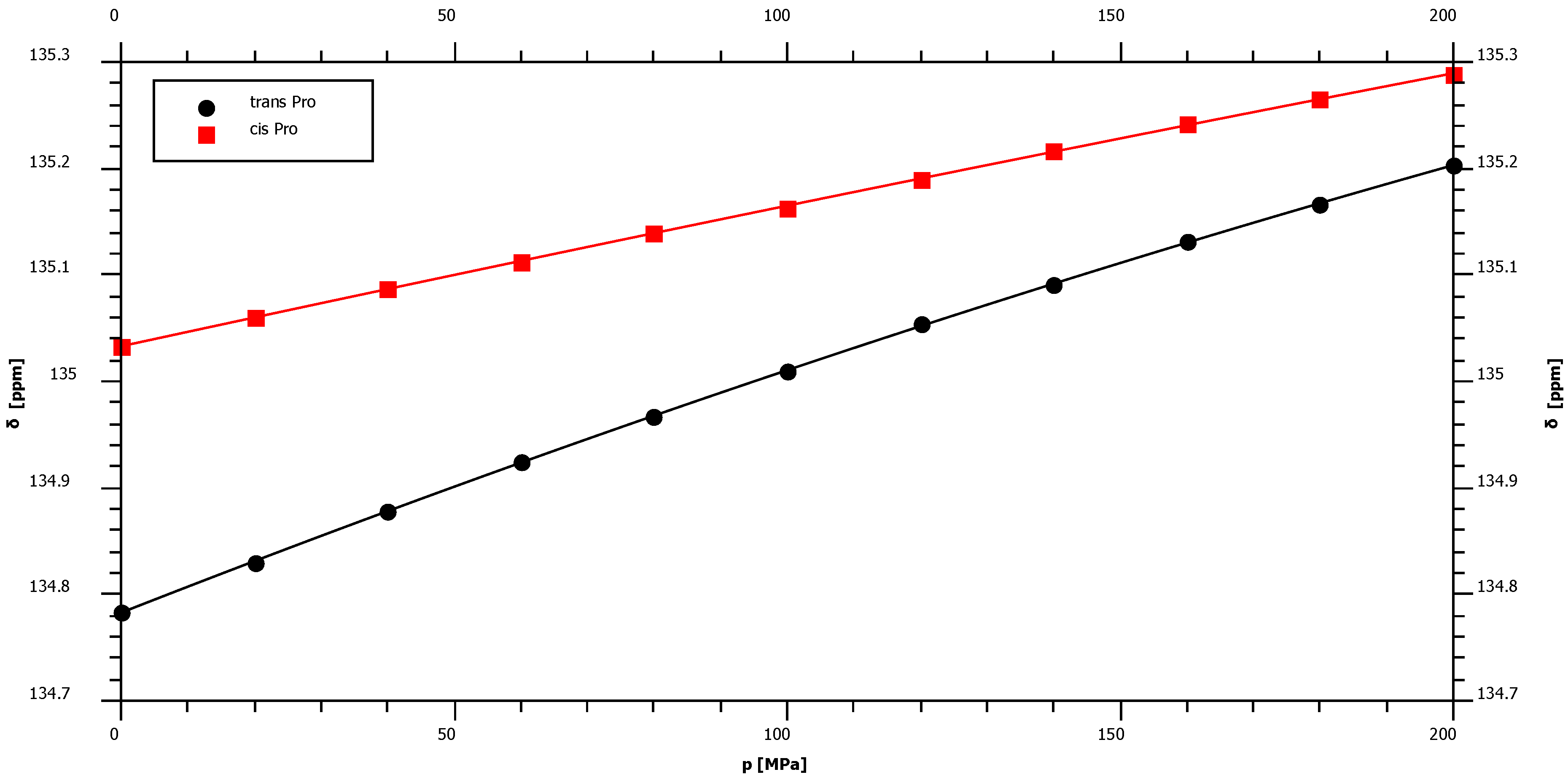
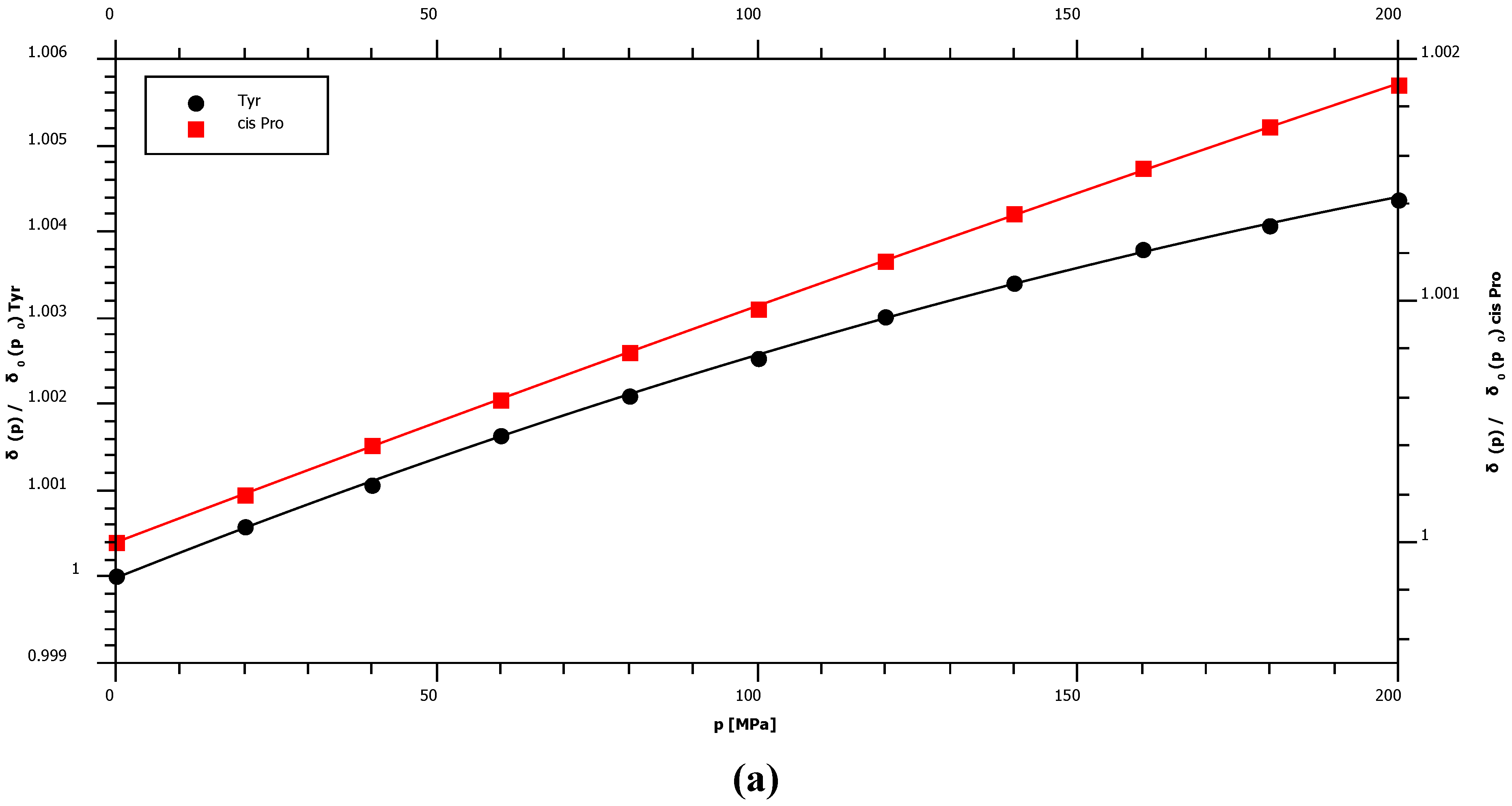
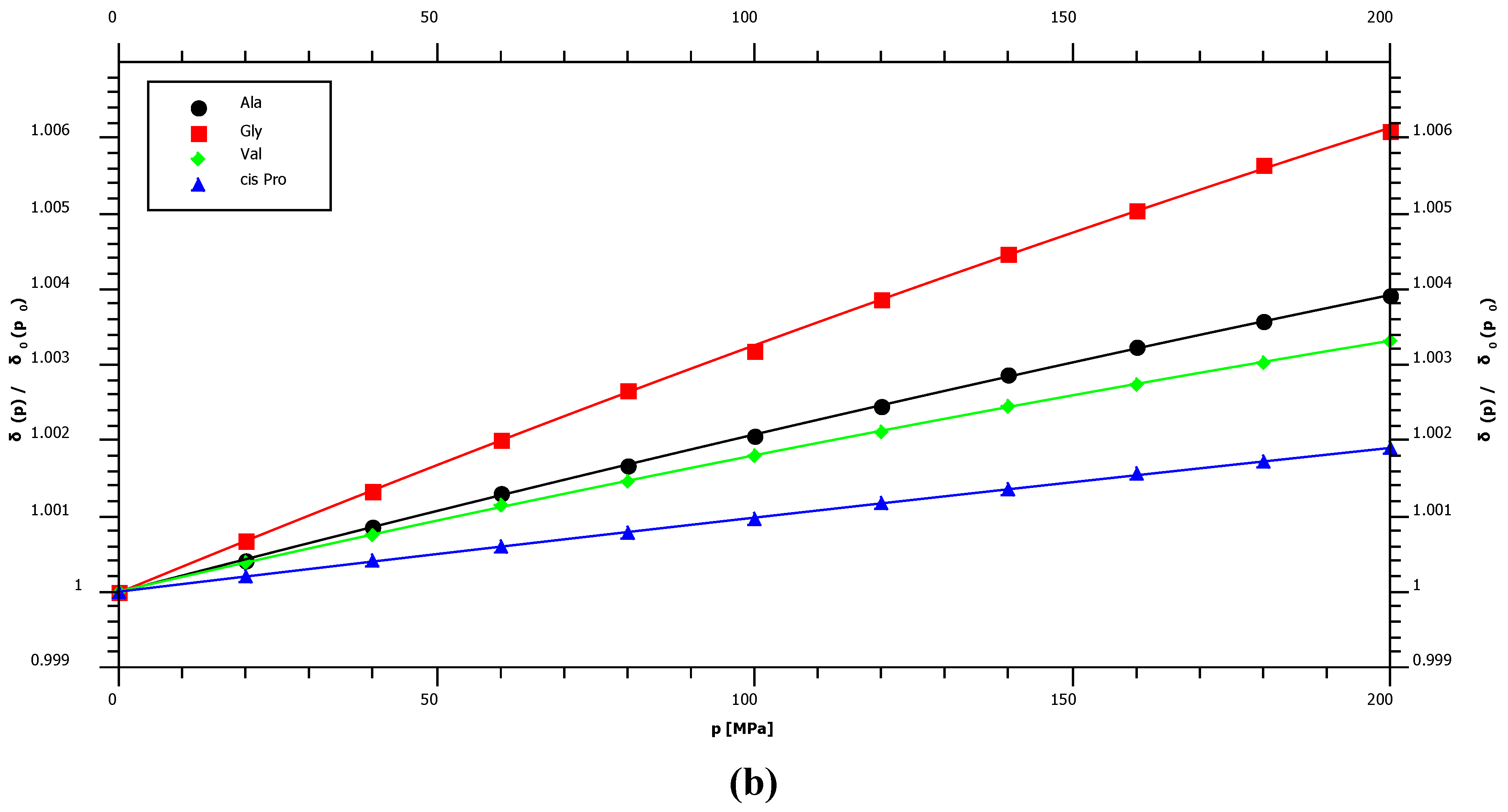
2.1. Pressure Dependence of the Backbone 15N Chemical Shifts
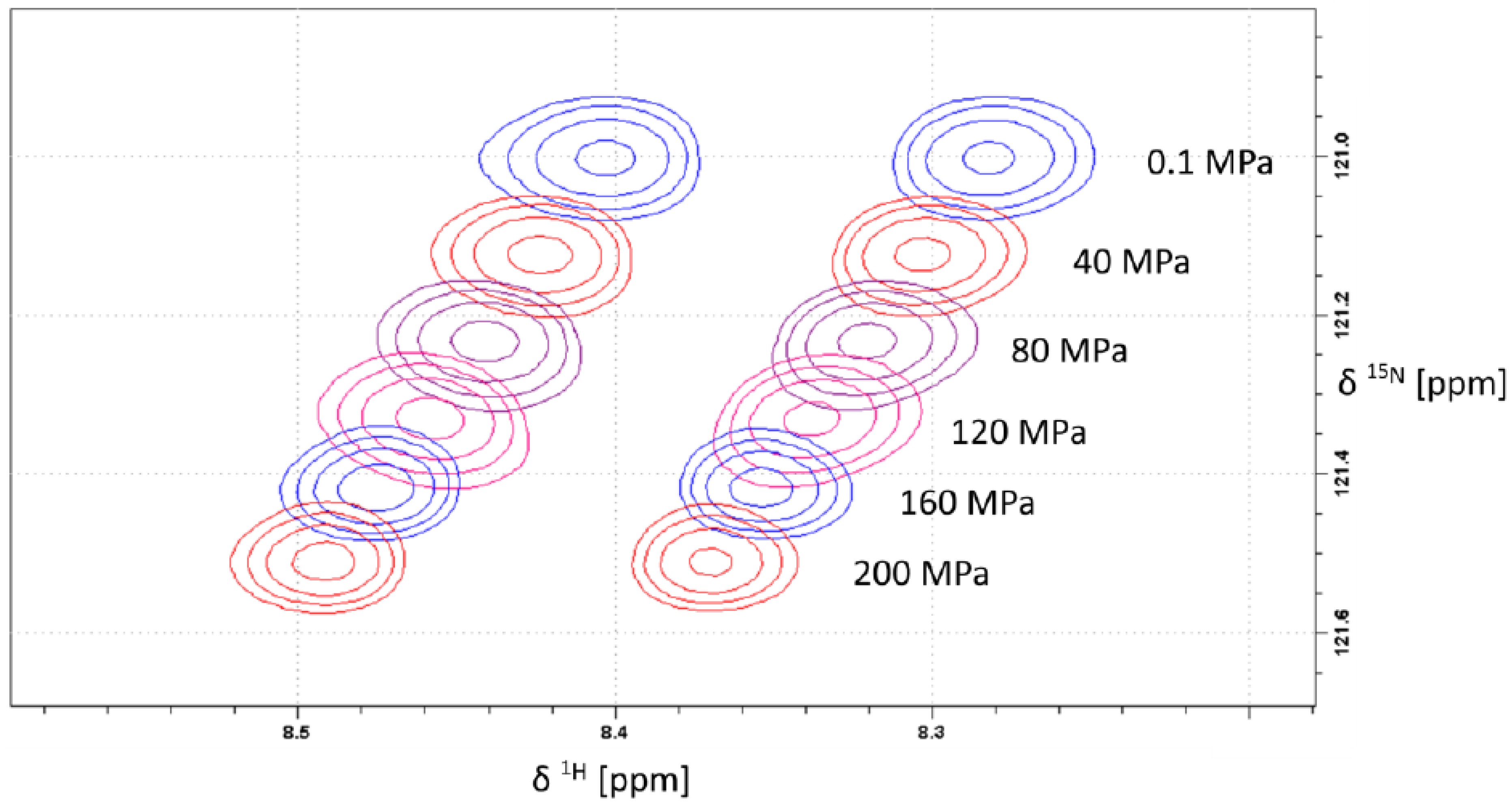
| δ015N | B115N | B215N | 1J1H15N | |||||
|---|---|---|---|---|---|---|---|---|
| (ppm) | (ppm/GPa) | (ppm/GPa2) | (Hz) | |||||
| Ala | 124.39 | 2.74 | ± | 0.03 | −1.49 | ± | 0.16 | −97.5 |
| Arg | 121.00 | 3.01 | ± | 0.04 | −2.44 | ± | 0.21 | −95.5 |
| Asn | 119.12 | 2.98 | ± | 0.17 | −1.73 | ± | 0.82 | −95.9 |
| Asp | 120.94 | 2.94 | ± | 0.07 | −2.58 | ± | 0.36 | −96.0 |
| Cys | 119.21 | 2.75 | ± | 0.08 | −1.87 | ± | 0.41 | −96.0 |
| Gln | 120.25 | 2.87 | ± | 0.06 | −2.15 | ± | 0.29 | −93.5 |
| Glu | 120.95 | 2.78 | ± | 0.17 | −1.66 | ± | 0.83 | −99.5 |
| Gly | 109.22 | 3.79 | ± | 0.08 | −2.16 | ± | 0.37 | −96.9 |
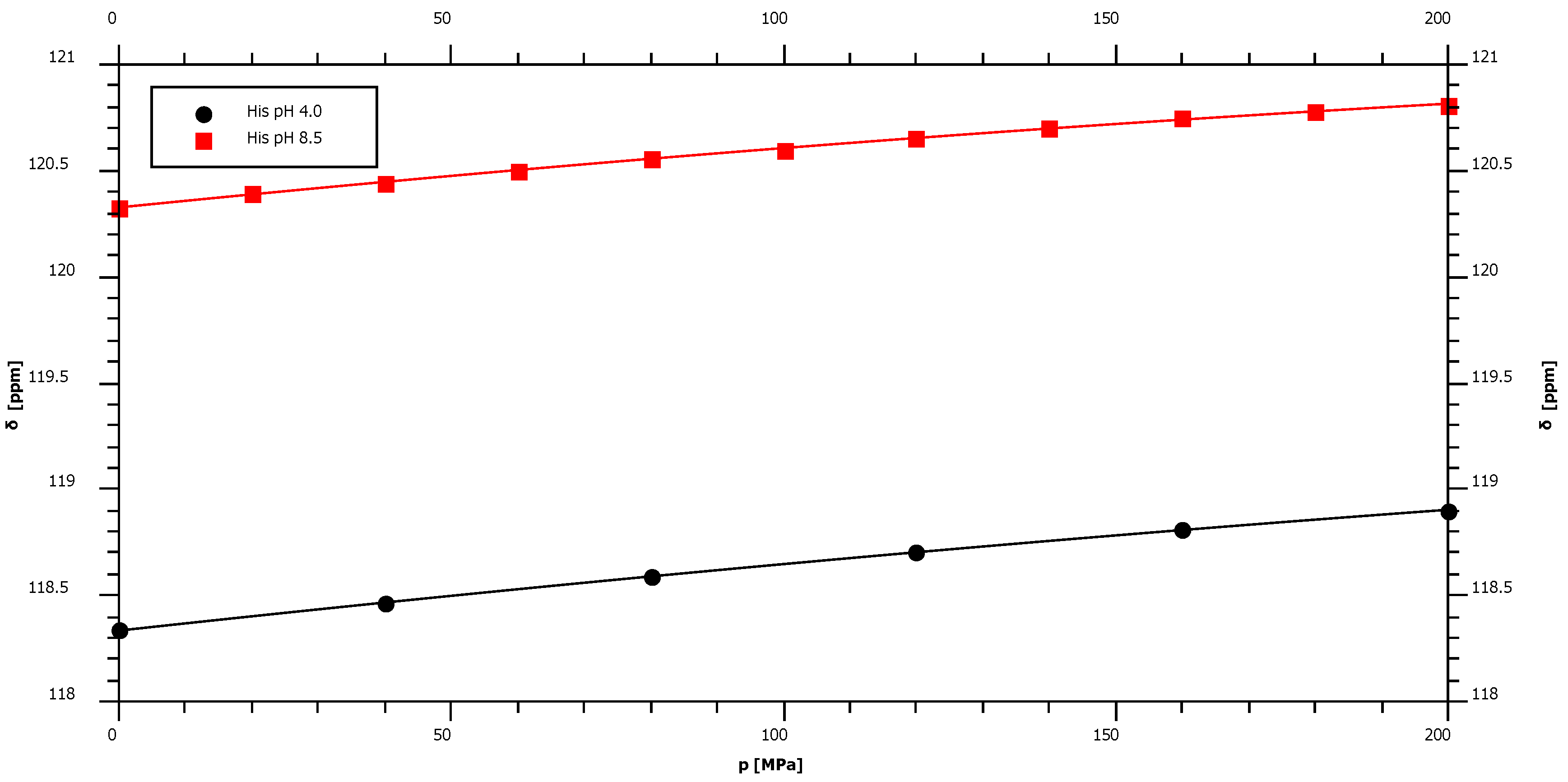
| His pH 4.0 | 118.34 | 3.42 | ± | 0.08 | −2.86 | ± | 0.38 | −98.2 |
| His pH 8.5* | 120.33 | 3.15 | ± | 0.10 | −3.53 | ± | 0.50 | – |
| Ile | 120.39 | 2.35 | ± | 0.03 | −1.89 | ± | 0.17 | −92.9 |
| Leu | 122.07 | 2.65 | ± | 0.10 | −2.92 | ± | 0.48 | −96.2 |
| Lys | 121.29 | 3.00 | ± | 0.03 | −2.24 | ± | 0.13 | −94.7 |
| Met | 120.17 | 2.76 | ± | 0.05 | −1.89 | ± | 0.22 | −92.7 |
| Phe | 120.62 | 3.01 | ± | 0.04 | −3.65 | ± | 0.19 | −95.5 |
| Pro trans* | 134.78 | 2.45 | ± | 0.02 | −1.76 | ± | 0.10 | – |
| Pro cis* | 135.03 | 1.36 | ± | 0.02 | −0.38 | ± | 0.10 | – |
| Ser | 116.27 | 3.41 | ± | 0.15 | −2.46 | ± | 0.70 | −95.7 |
| Thr | 114.14 | 3.28 | ± | 0.05 | −2.36 | ± | 0.24 | −93.8 |
| Trp | 121.43 | 3.35 | ± | 0.03 | −2.90 | ± | 0.17 | −95.5 |
| Tyr | 120.33 | 3.56 | ± | 0.07 | −4.52 | ± | 0.32 | −93.9 |
| Val | 119.41 | 2.32 | ± | 0.02 | −1.70 | ± | 0.12 | −95.0 |
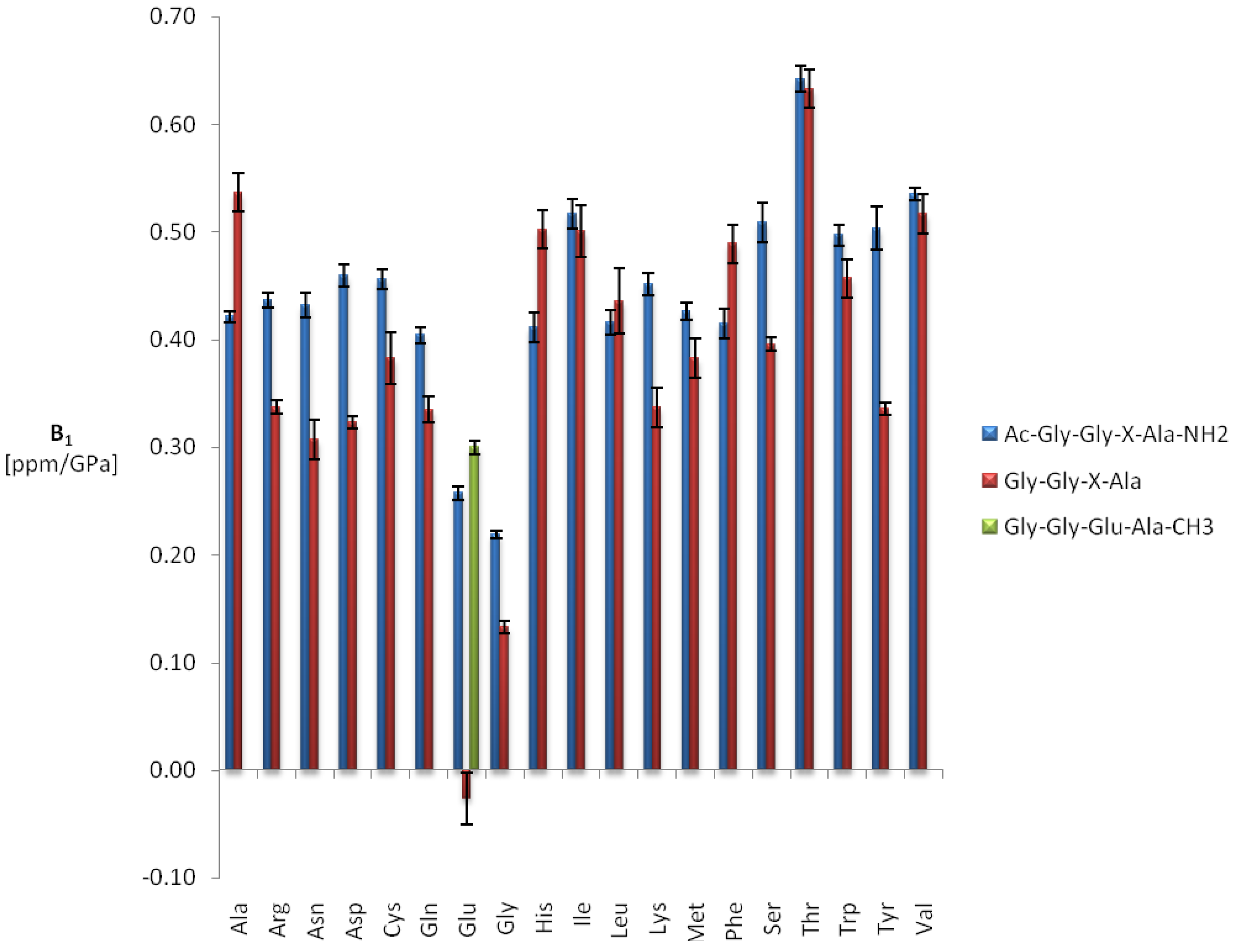
2.2. Measurement of the One-Bond Amide Coupling Constants
2.3. Pressure Dependence of the Amide 1H Chemical Shifts
| δ0HN | B1HN | B2HN | |||||
|---|---|---|---|---|---|---|---|
| (ppm) | (ppm/GPa) | (ppm/GPa2) | |||||
| Ala | 8.368 | 0.45 | ± | 0.02 | −0.15 | ± | 0.08 |
| Arg | 8.342 | 0.48 | ± | 0.02 | −0.22 | ± | 0.10 |
| Asn | 8.503 | 0.50 | ± | 0.03 | −0.42 | ± | 0.14 |
| Asp | 8.433 | 0.56 | ± | 0.02 | −0.48 | ± | 0.09 |
| Cys | 8.418 | 0.55 | ± | 0.01 | −0.49 | ± | 0.05 |
| Gln | 8.433 | 0.44 | ± | 0.03 | −0.17 | ± | 0.13 |
| Glu | 8.539 | 0.25 | ± | 0.02 | −0.03 | ± | 0.08 |
| Gly | 8.449 | 0.24 | ± | 0.01 | −0.10 | ± | 0.06 |
| His pH 4.0 | 8.484 | 0.51 | ± | 0.01 | −0.47 | ± | 0.06 |
| Ile | 8.154 | 0.64 | ± | 0.03 | −0.64 | ± | 0.14 |
| Leu | 8.279 | 0.52 | ± | 0.02 | −0.50 | ± | 0.12 |
| Lys | 8.317 | 0.55 | ± | 0.02 | −0.49 | ± | 0.10 |
| Met | 8.398 | 0.49 | ± | 0.02 | −0.31 | ± | 0.10 |
| Phe | 8.263 | 0.55 | ± | 0.02 | −0.68 | ± | 0.10 |
| Ser | 8.434 | 0.64 | ± | 0.05 | −0.65 | ± | 0.26 |
| Thr | 8.269 | 0.75 | ± | 0.02 | −0.56 | ± | 0.11 |
| Trp | 8.129 | 0.58 | ± | 0.02 | −0.42 | ± | 0.11 |
| Tyr | 8.190 | 0.67 | ± | 0.05 | −0.82 | ± | 0.25 |
| Val | 8.232 | 0.58 | ± | 0.02 | −0.21 | ± | 0.08 |
2.4. pH-Dependence of the Pressure Response
3. Experimental Section
3.1. Synthesis of Peptides
3.2. Sample Preparation
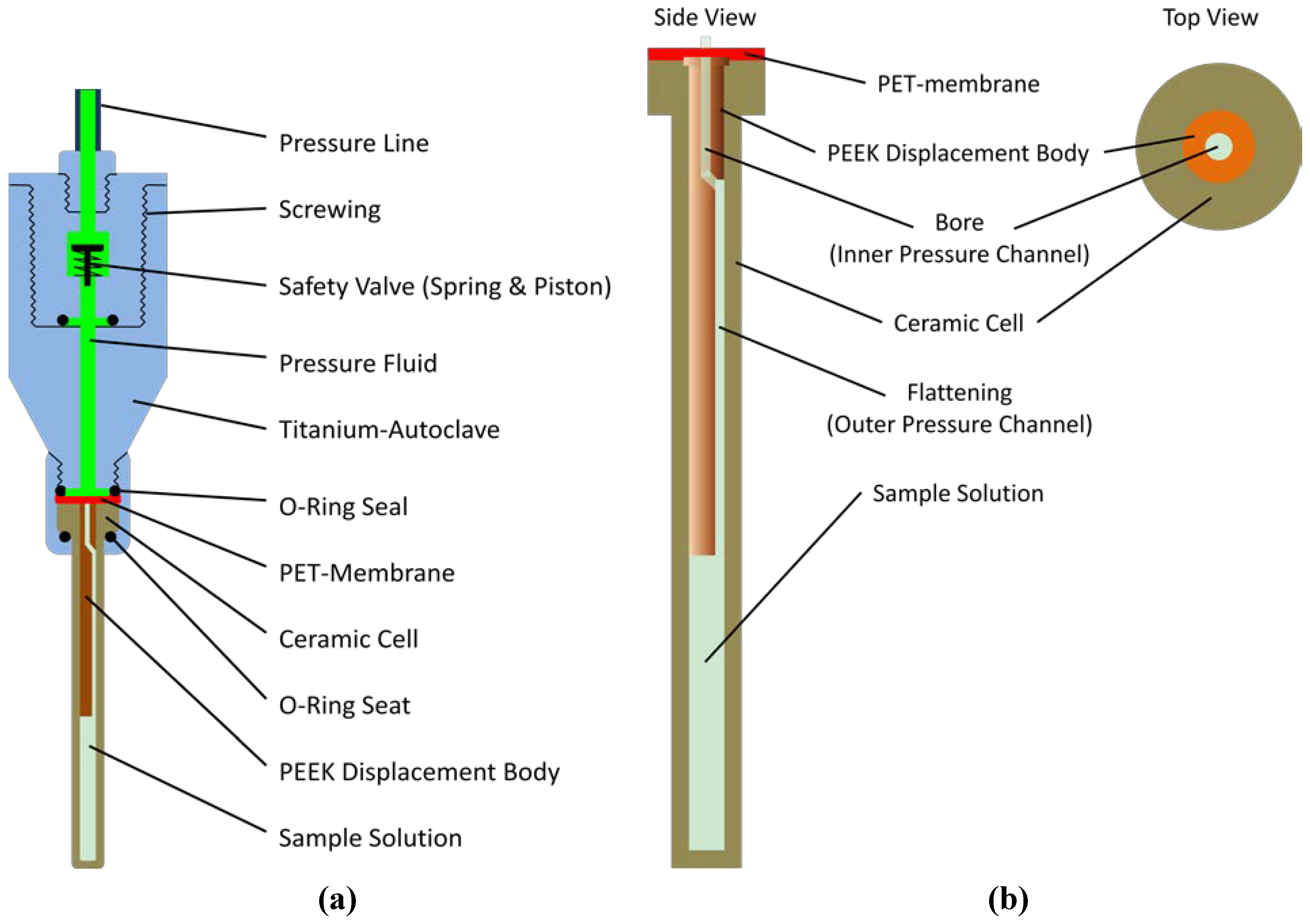
3.3. High Pressure System
3.4. NMR Spectroscopy
3.5. Data Evaluation
4. Conclusions
Acknowledgements
References
- Akasaka, K. Probing conformational fluctuations by pressure perturbations. Chem. Rev. 2006, 106, 1814–1835. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yamada, H.; Akasaka, K. Effect of pressure on individual hydrogen bonds in proteins. Basic pancreatic trypsin inhibitor. Biochemistry 1998, 37, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Kalbitzer, H.R.; Görler, A.; Li, H. 15N and 1H NMR study of Histidine containing protein (HPr) from Staphylococcus carnosus at high-pressure. Protein Sci. 2000, 9, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, A.J.; Kay, L.E. NMR spectroscopy brings invisible protein states into focus. Nat. Chem. Biol. 2009, 11, 808–814. [Google Scholar] [CrossRef]
- Arnold, M.A.; Kremer, W.; Luedemann, H.D.; Kalbitzer, H.R. 1H-NMR parameters of common amino acid residues measured in aqueous solutions of the linear tetrapeptides Gly-Gly-X-Ala at pressures between 0.1 and 200 MPa. Biophys. Chem. 2002, 96, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Kremer, W.; Kachel, N.; Kuwata, K.; Akasaka, K.; Kalbitzer, H.R. Species specific differences in the intermediate states of human and Syrian hamster prion protein detected by high pressure NMR spectroscopy. J. Biol. Chem. 2007, 282, 22689–22698. [Google Scholar] [CrossRef] [PubMed]
- Bundi, A.; Wuethrich, K. 1H-NMR parameters of the common amino acid residues measured in aqueous solutions of the linear tetrapeptides H-Gly-Gly-X-L-Ala-OH. Biopolymers 1979, 18, 285–297. [Google Scholar] [CrossRef]
- Asakawa, N.; Kameda, T.; Kuroki, S.; Kurosu, H.; Ando, S.; Shoji, A. Structural studies of hydrogen-bonded peptides and polypeptides by solid state. Ann. Reports NMR Spectr. 1998, 35, 56–137. [Google Scholar]
- Kjaergaard, M.; Brander, S.; Poulsen, F.M. Random coil chemical shift for intrinsically disordered proteins: Effects of temperature and pH. J. Biomol. NMR 2011, 49, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Kremer, W. High-pressure NMR studies in proteins. Ann. Reports NMR Spectr. 2006, 57, 177–203. [Google Scholar]
- Bundi, A.; Wuethrich, K. Use of amide 1H-NMR titration shifts for studies of polypeptide conformation. Biopolymers 1979, 18, 299–311. [Google Scholar] [CrossRef]
- Quinlan, R.J.; Reinhart, G.D. Baroresistant buffer mixtures for biochemical analyses. Anal. Biochem. 2005, 341, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Huberth, A. Hochdruck-NMR am Ras-Protein. Diploma thesis in Physics, University of Regensburg, July 2011. [Google Scholar]
- Peterson, R.W.; Wand, A.J. Self-contained high-pressure cell, apparatus, and procedure for the preparation of encapsulated proteins dissolved in low viscosity fluids for nuclear magnetic resonance spectroscopy. Rev. Sci. Instrum. 2005, 76, 094101:1–094101:7. [Google Scholar] [CrossRef]
- Erlach, M.B.; Munte, C.E.; Kremer, W.; Hartl, R.; Rochelt, D.; Niesner, D.; Kalbitzer, H.R. Ceramic cells for high pressure NMR spectroscopy on proteins. J. Magn. Reson. 2010, 204, 196–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, H. Pressure-resisting glass cell for high pressure, high resolution NMR measurement. Rev. Sci. Instrum. 1974, 45, 640–642. [Google Scholar] [CrossRef]
- Raiford, D.S.; Fisk, C.L.; Becker, E.D. Calibration of methanol and ethylene glycol nuclear magnetic resonance thermometers. Anal. Chem. 1979, 51, 2050–2051. [Google Scholar] [CrossRef]
- Davis, A.L.; Keeler, J.; Laue, E.D.; Moskau, D. Experiments for recording pure-absorption heteronuclear correlation spectra using pulsed field gradients. J. Magn. Reson. 1992, 98, 207–216. [Google Scholar]
- Kanelisa, V.; Donaldson, L.; Muhandiram, D.R.; Rotin, D.; Forman-Kay, J.D.; Kay, L.E. Sequential assignment of proline-rich regions in proteins: Application to modular binding domain complexes. J. Biomol. NMR 2000, 16, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.C.; Grzesiek, S.; Tschudin, R.; Lodi, P.J.; Bax, A. Sequential backbone assignment of isotopically enriched proteins in D2O by deuterium-decoupled HA(CA)N and HA(CACO)N. J. Biomol. NMR 1995, 5, 376–382. [Google Scholar] [PubMed]
- Simpson, A.J.; Brown, S.A. Purge NMR: Effective and easy solvent suppression. J. Magn. Reson. 2005, 175, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Bigam, C.G.; Yao, J.; Abildgaard, F.; Dyson, H.J.; Oldfield, E.; Markley, J.L.; Sykes, B.D. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 1995, 6, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Kachel, N.; Kremer, W.; Zahn, R.; Kalbitzer, H.R. Observation of intermediate states of the human prion protein by high pressure NMR spectroscopy. BMC Struct. Biol. 2006, 6, 16:1–16:18. [Google Scholar] [CrossRef]
© 2012 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Koehler, J.; Beck Erlach, M.; Crusca, E.; Kremer, W.; Munte, C.E.; Kalbitzer, H.R. Pressure Dependence of 15N Chemical Shifts in Model Peptides Ac-Gly-Gly-X-Ala-NH2. Materials 2012, 5, 1774-1786. https://doi.org/10.3390/ma5101774
Koehler J, Beck Erlach M, Crusca E, Kremer W, Munte CE, Kalbitzer HR. Pressure Dependence of 15N Chemical Shifts in Model Peptides Ac-Gly-Gly-X-Ala-NH2. Materials. 2012; 5(10):1774-1786. https://doi.org/10.3390/ma5101774
Chicago/Turabian StyleKoehler, Joerg, Markus Beck Erlach, Edson Crusca, Werner Kremer, Claudia E. Munte, and Hans Robert Kalbitzer. 2012. "Pressure Dependence of 15N Chemical Shifts in Model Peptides Ac-Gly-Gly-X-Ala-NH2" Materials 5, no. 10: 1774-1786. https://doi.org/10.3390/ma5101774




