A Genetic Algorithm Using Triplet Nucleotide Encoding and DNA Reproduction Operations for Unconstrained Optimization Problems
Abstract
:1. Introduction
- We define a new DNA coding scheme which encodes the potential solution problem space using triplet nucleotides that represent amino acids.
- We define a set of evolutional operations that create new individuals in the problem space by mimicking the DNA reproduction process at an amino acid level.
- We present a genetic algorithm that uses triplet nucleotide encoding (TNE) and a DNA reproduction operator (DRO), hence the name GA-TNE+DRO.
- We perform experiments to evaluate the performance of the algorithm using a benchmark of eight unconstrained optimization problems and compare it with state-of-the-art algorithms including conventional GA [1], PSO [2], and DE [3]. Our experimental results show that our algorithm can converge to solutions much closer to the global optimal solutions in a much lower number of iterations than the existing algorithms.
2. A Triplet Nucleotide Coding Scheme
3. A Set of DNA Reproduction Operations
3.1. Crossover Operations
- (1)
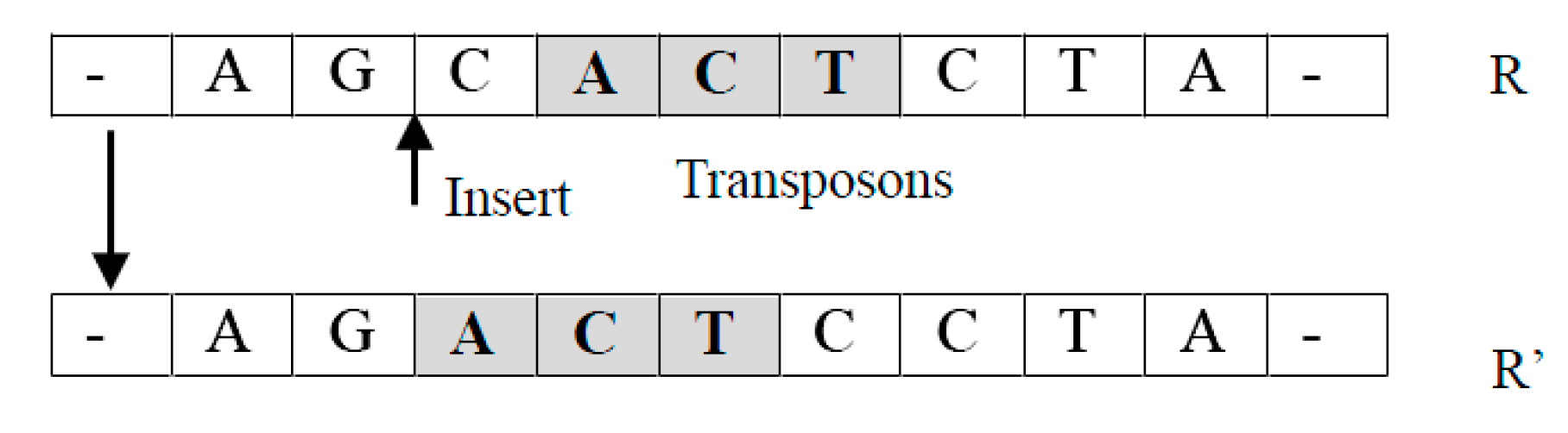
- Translocation operator TransLoc(R): It takes an individual as an input and returns a new individual by relocating a randomly selected unit of to a randomly chosen new location.For example, suppose , where is a unit. A new individual returned can be in which has been moved into the position before . Notice that it is easy to extend this operation so that an arbitrary new location can be selected (See Figure 2).
- (2)
- Transformation operator Transform(R): It takes an individual and two positions as parameters, and returns a new individual by swapping two randomly selected units.For example, the sequence becomes , after exchanging randomly selected units with .
- (3)
- Permutation operator Permute(R): It takes an individual as a parameter, and returns a new individual in which a randomly selected unit is randomly permuted.For example, if , the new individual can be , where is a random permutation of a randomly selected unit .
3.2. Mutation Operations
- (1)
- Inverse anticodon mutation IA(R). It takes an individual as a parameter and returns a new individual by replacing a randomly selected unit (as a codon) with its inverse anticodon. In biology, the anticodon of a codon is obtained by replacing each nucleotide with its complementary nucleotide based on the Watson–Crick complementary principle. Thus, A is replaced by T, C by G, and vice versa. An inversed anticodon is obtained by inverting the nucleotide sequence of the anticodon.For example, if the randomly selected codon is GCA (or 112 in numerical code), its anticodon will be CCT (or 003 in coding) and its inverse anticodon is TCC (or 300 in coding).
- (2)
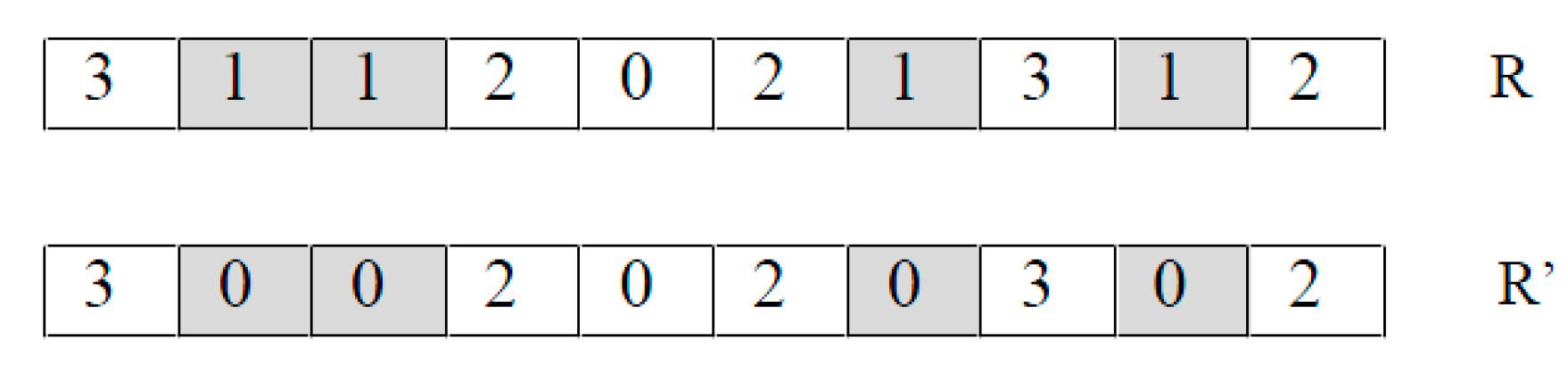
- Frequency mutation FM(R). It takes an individual as a parameter and returns a new individual by replacing every occurrence of the most frequently appearing nucleotide by the least frequently appearing nucleotide.For example, in Figure 3 nucleotide G (represented by 1) is the most frequently appearing and nucleotide C (represented by 0) is the least frequently appearing. Thus, the FM operator replaces every G using a C.
- (3)
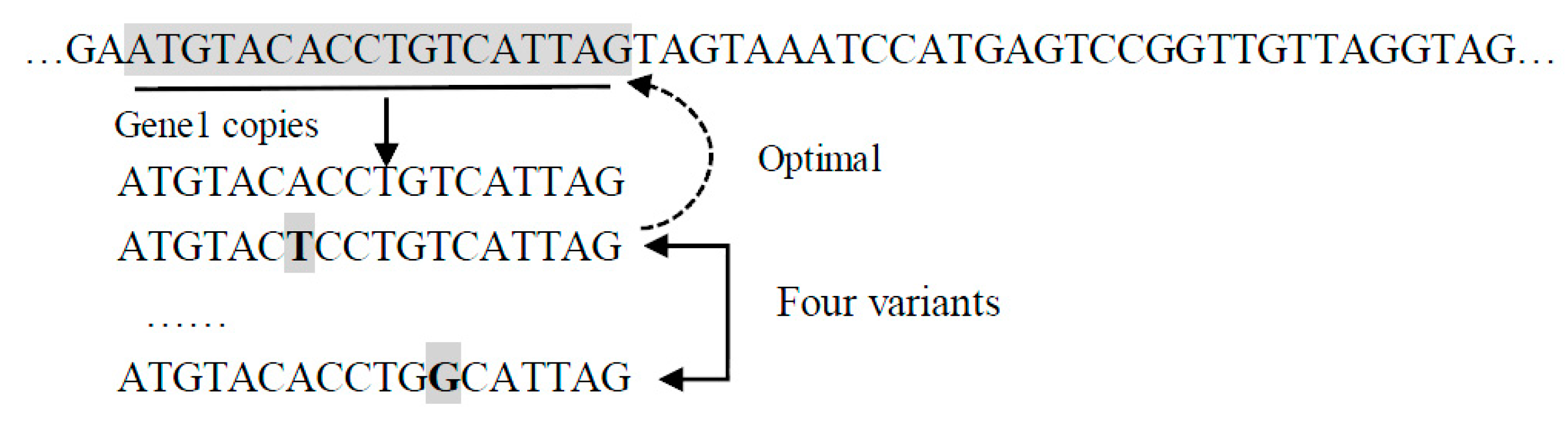
- Pseudo-bacteria mutation Pseudobac(R). It takes an individual as a parameter and returns a new individual as follows.First, a random subsequence of nucleotides is identified as a gene. Then, a given number of candidate individuals are created by randomly changing one nucleotide in the selected gene. Finally, the candidate individual with the highest fitness value is returned.For example, in Figure 4, gene1 is randomly selected, and five variations are created by changing one nucleotide at a time (indicated by the shaded bold letter). One candidate individual is created by using each of the variants to replace gene1.
3.3. Recombination Operation

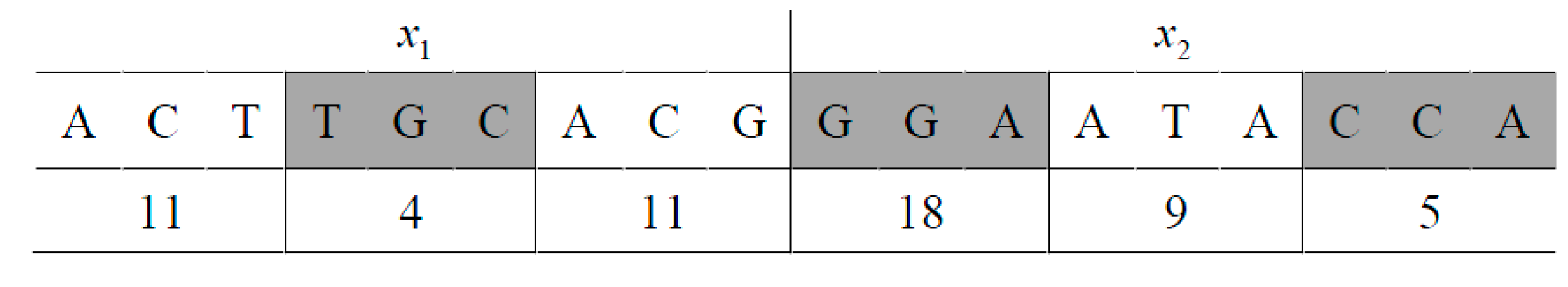
 respectively. The restriction enzymes TaqI and SciNI will cleave the above two molecules R1 and R2 into four segments:
respectively. The restriction enzymes TaqI and SciNI will cleave the above two molecules R1 and R2 into four segments:

4. The GA-TNE+DRO Algorithm
| Algorithm 1 GA-TNE+DRO |
| Input: : the objective function : the number of variables in : domains of the n variables : the size of initial population Pc: the probability of crossover operation Pm: the probability of mutation operation Pr: the probability of recombination operation : max number of iterations : accuracy threshold Output: The value of X that optimizes Method: 1. POP = a population of randomly generated individuals 2. For each p in POP 3. Calculate the fitness value of p using f(decode(p)) 4. = the best individual in POP 5. OldX = any individual in POP that is not 6. While termination condition is not satisfied do 7. OldX = 8. NewPOP = {} 9. NEU = {N/2 neutral individuals in POP} 10. DEL = {N/2 deleterious individuals in POP} 11. For each individual p in NEU 12. Apply crossover operations to p according to Pc and add results to NewPOP 13. For each individual p in DEL 14. Apply mutation operations to p according to Pm and add results to NewPOP 15. For each pair of individual p and p’ in POP 16. Apply the recombination operation according to Pr and add results to NewPOP 17. For each p in NewPOP 18. Calculate the fitness value of p using f(decode(p)) 19. = the best individual in NewPOP 20. If is not better than OldX then 21. Generate randomly new individuals and pick up the best one into NewPOP 22. POP = NewPOP 23. End while 24. Return decode (X) |
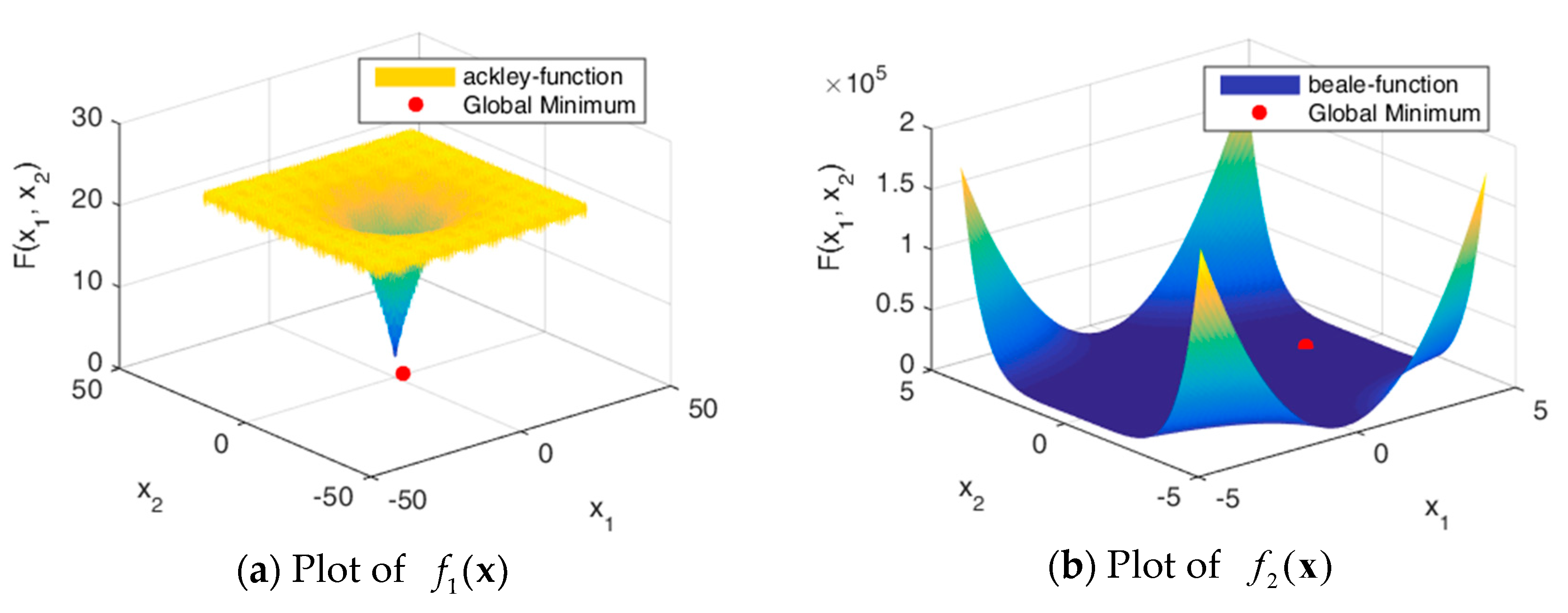
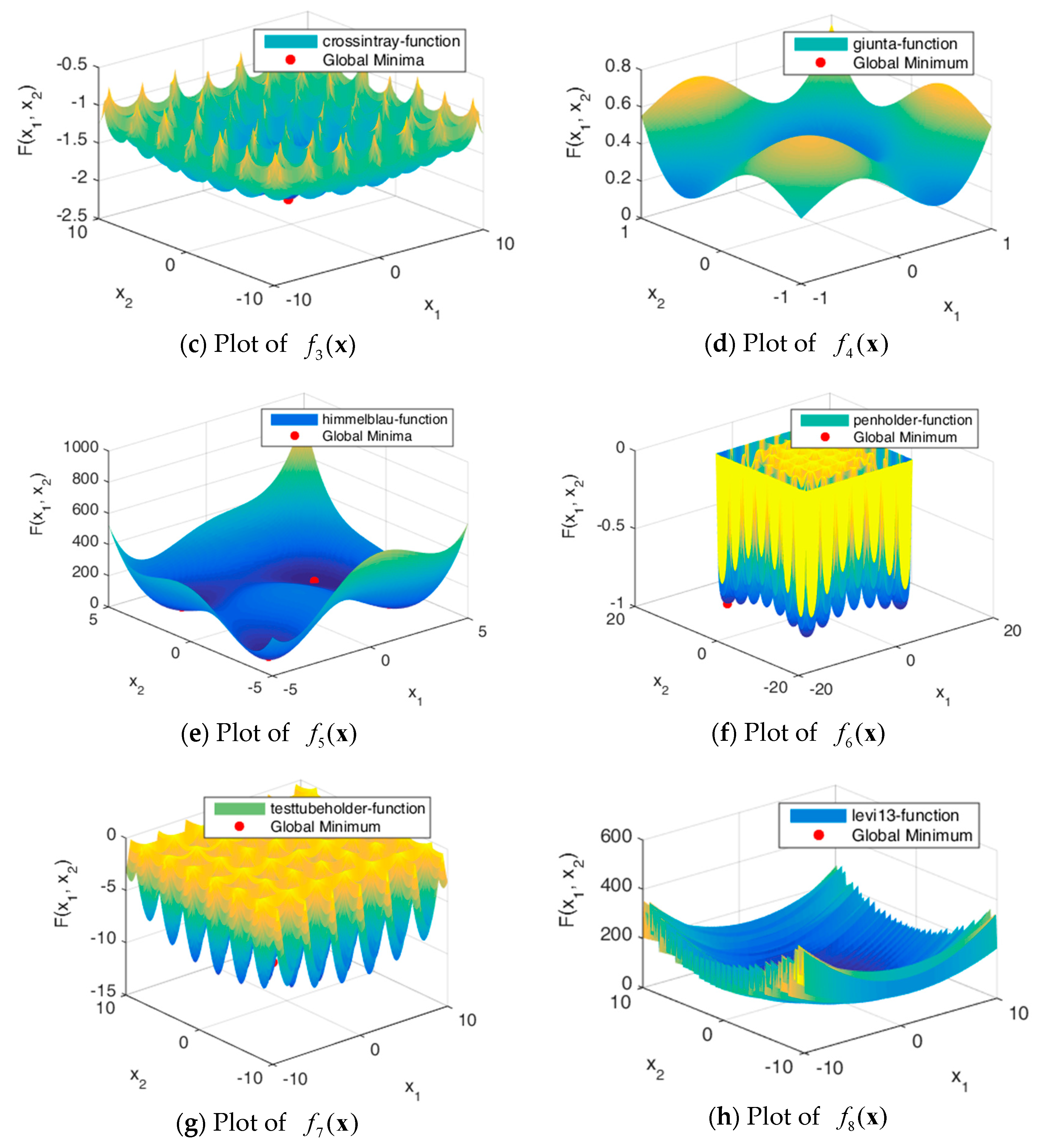
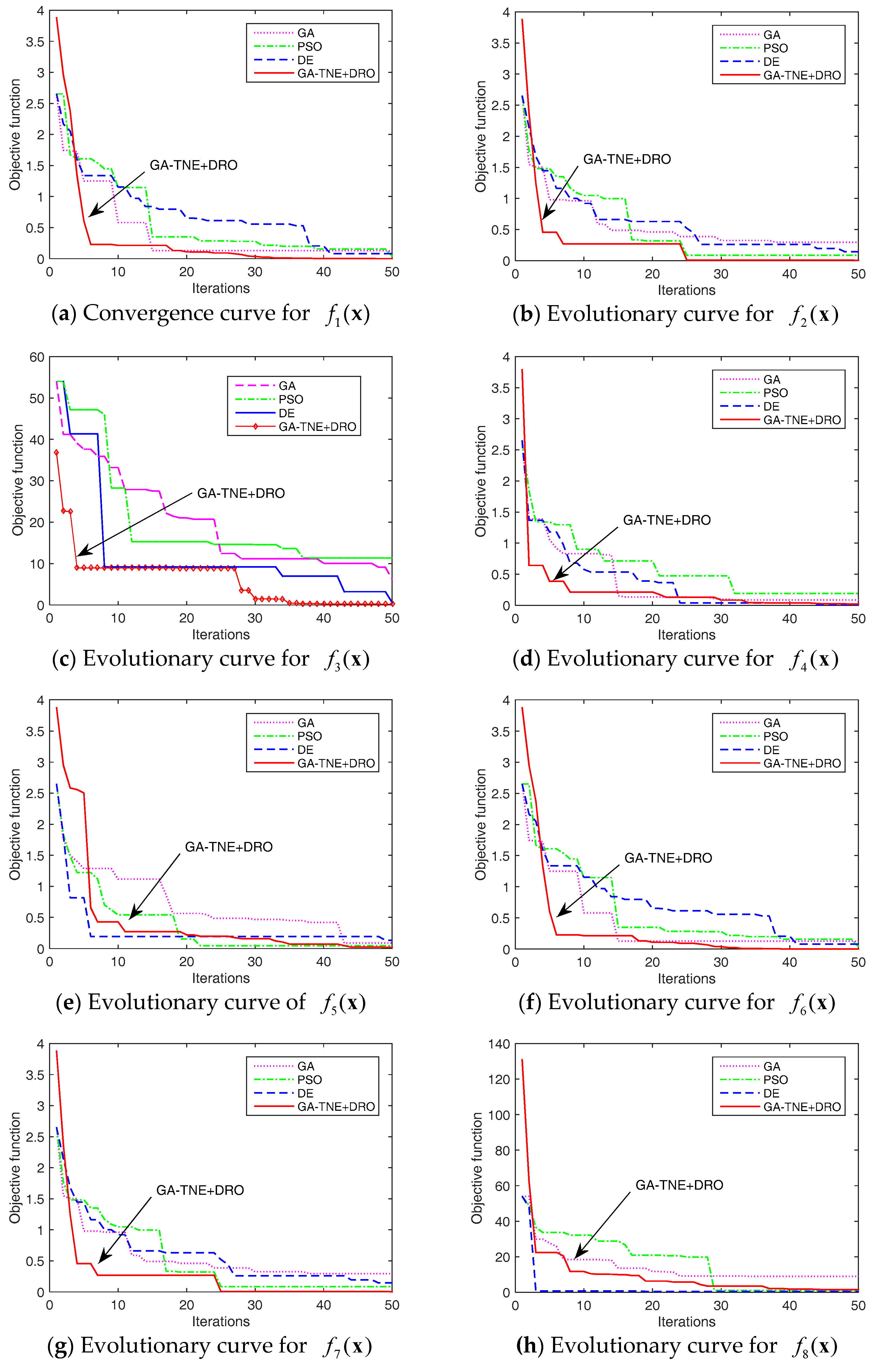
5. Numerical Experiments
5.1. Experiment Setup
5.2. Results and Discussion
5.3. Algorithm Complexity
| Algorithm 2 Test Problem |
| for i = 1:200 x = 0.55 + i; x = x + x; x = x/ 2; x = x*x; x= sqrt(x); x = log(x); x= exp(x); x = x/(x+2); end |
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Holland, J.H. Adaptation in Natural and Artificial Systems; University of Michigan Press: Ann Arbor, MI, USA, 1975. [Google Scholar]
- Guedria, N.B. Improved accelerated PSO algorithm for mechanical engineering optimization problems. Appl. Soft Comput. 2016, 40, 455–467. [Google Scholar] [CrossRef]
- Storn, R.; Price, K. Differential Evolution—A simple and efficient heuristic for global optimization over continuous spaces. J. Glob. Optim. 1997, 11, 341–359. [Google Scholar] [CrossRef]
- Garg, H. Solving structural engineering design optimization problems using an artificial bee colony algorithm. J. Ind. Manag. Optim. 2014, 10, 777–794. [Google Scholar] [CrossRef]
- Garg, H.; Sharma, S.P. Multi-objective reliability redundancy allocation problem using particle swarm optimization. Comput. Ind. Eng. 2013, 64, 247–255. [Google Scholar] [CrossRef]
- Fan, Q.; Yan, X. Self-adaptive differential evolution algorithm with discrete mutation control parameters. Expert Syst. Appl. 2015, 42, 1551–1572. [Google Scholar] [CrossRef]
- Garg, H. A hybrid GA-GSA algorithm for optimizing the performance of an industrial system by utilizing uncertain data. In Handbook of Research on Artificial Intelligence Techniques and Algorithms; IGI Global: Hershey, PA, USA, 2015; pp. 620–654. [Google Scholar]
- Pelusi, D.; Mascella, R.; Tallini, L. Revised gravitational search algorithms based on evolutionary-fuzzy systems. Algorithms 2017, 10, 44. [Google Scholar] [CrossRef]
- Garg, H. A hybrid PSO-GA algorithm for constrained optimization problems. Appl. Math. Comput. 2016, 274, 292–305. [Google Scholar] [CrossRef]
- Garg, H. An efficient biogeography based optimization algorithm for solving reliability optimization problems. Swarm Evol. Comput. 2015, 24, 1–10. [Google Scholar] [CrossRef]
- Adleman, L.M. Molecular computation of solutions to combinatorial problems. Science 1994, 266, 1021–1024. [Google Scholar] [CrossRef] [PubMed]
- Nabil, B.; Guenda, K.; Gulliver, A. Construction of codes for DNA computing by the greedy algorithm. ACM Commun. Comput. Algebra. 2015, 49. [Google Scholar] [CrossRef]
- Mayukh, S.; Ghosal, P. Implementing Data Structure Using DNA: An Alternative in Post CMOS Computing. In Proceedings of the 2015 IEEE Computer Society Annual Symposium on VLSI, Montpellier, France, 8–10 July 2015. [Google Scholar]
- Huang, Y.; Tian, Y.; Yin, Z. Design of PID controller based on DNA COMPUTING. In Proceedings of the International Conference on Artificial Intelligence and Computational Intelligence (AICI), Sanya, China, 23–24 October 2010; pp. 195–198. [Google Scholar]
- Yongjie, L.; Jie, L. A feasible solution to the beam-angle-optimization problem in radiotherapy planning with a DNA-based genetic algorithm. IEEE Trans. Biomed. Eng. 2010, 57, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Damm, R.B.; Resende, M.G.; Débora, P.R. A biased random key genetic algorithm for the field technician scheduling problem. Comput. Oper. Res. 2016, 75, 49–63. [Google Scholar] [CrossRef]
- Li, H.B.; Demeulemeester, E. A genetic algorithm for the robust resource leveling problem. J. Sched. 2016, 19, 43–60. [Google Scholar] [CrossRef]
- Ding, Y.S.; Ren, L.H. DNA genetic algorithm for design of the generalized membership-type Takagi-Sugeno fuzzy control system. In Proceedings of the 2000 IEEE International Conference on Systems, Man, and Cybernetics, Nashville, TN, USA, 8–11 October 2000; pp. 3862–3867. [Google Scholar]
- Zhang, L.; Wang, N. A modified DNA genetic algorithm for parameter estimation of the 2-chlorophenol oxidation in supercritical water. Appl. Math. Model. 2013, 37, 1137–1146. [Google Scholar] [CrossRef]
- Chen, X.; Wang, N. Optimization of short-time gasoline blending scheduling problem with a DNA based hybrid genetic algorithm. Comput. Chem. Eng. 2010, 49, 1076–1083. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, N. An adaptive RNA genetic algorithm for modeling of proton exchange membrane fuel cells. Int. J. Hydrog. Energy 2013, 38, 219–228. [Google Scholar] [CrossRef]
- Zang, W.K.; Sun, M.H.; Jiang, Z.N. A DNA genetic algorithm inspired by biological membrane structure. J. Comput. Theor. Nanosci. 2016, 13, 3763–3772. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, N.; Bi, Y. Type-1/type-2 fuzzy logic systems optimization with RNA genetic algorithm for double inverted pendulum. Appl. Math. Model. 2015, 39, 70–85. [Google Scholar] [CrossRef]
- Zang, W.K.; Ren, L.Y.; Zhang, W.Q.; Liu, X.Y. Automatic density peaks clustering using DNA genetic algorithm optimized data field and Gaussian process. Int. J. Pattern Recognit. Artif. Intell. 2017, 31, 1750023. [Google Scholar] [CrossRef]
- Zang, W.K.; Jiang, Z.N.; Ren, L.Y. Spectral clustering based on density combined with DNA genetic algorithm. Int. J. Pattern Recognit. Artif. Intell. 2017, 31, 7799. [Google Scholar] [CrossRef]
- Zang, W.K.; Sun, M.H. Searching parameter values in support vector machines using DNA genetic algorithms. Lect. Notes Comput. Sci. 2016, 9567, 588–598. [Google Scholar]
- Yoshikawa, T.; Furuhashi, T.; Uchikawa, Y. The effects of combination of DNA coding method with pseudo-bacterial GA. In Proceedings of the 1997 IEEE International Conference on Evolutionary Computation, Indianapolis, IN, USA, 13–16 April 1997; pp. 285–290. [Google Scholar]
- Amos, M.; Păun, G.; Rozenberg, G.; Salomaa, A. Topics in the theory of DNA computing. Theor. Comput. Sci. 2002, 287, 3–38. [Google Scholar] [CrossRef]
- Cheng, W.; Shi, H.; Xin, X.; Li, D. An elitism strategy based genetic algorithm for streaming pattern discovery in wireless sensor networks. Commun. Lett. IEEE 2011, 15, 419–421. [Google Scholar] [CrossRef]
- Neuhauser, C.; Krone, S.M. The genealogy of samples in models with selection. Genetics 1997, 145, 519–534. [Google Scholar] [PubMed]
- Haupt, R.L.; Haupt, S.E. Practical Genetic Algorithms, 2nd ed.; John Wiley & Sons, Inc.: Camp Hill, PA, USA, 2004. [Google Scholar]
- Mirjalili, S.; Mirjalili, S.M.; Hatamlou, A. Multi-verse optimizer: A nature inspired algorithm for global optimization. Neural Comput. Appl. 2015. [Google Scholar] [CrossRef]
- Liang, J.; Qu, B.; Suganthan, P. Problem Definitions and Evaluation Criteria for the CEC 2014 Special Session and Competition on Single Objective Real-Parameter Numerical Optimization; Technical Report; Computational Intelligence Laboratory, Zhengzhou University: Zhengzhou, China; Nanyang Technological University: Singapore, 2013. [Google Scholar]
| First Nucleotide | Second Nucleotide | Third Nucleotide | |||
|---|---|---|---|---|---|
| T | C | A | G | ||
| T | Phe(0) | Ser(2) | Tyr(3) | Cys(4) | T |
| T | Phe(0) | Ser(2) | Tyr(3) | Cys(4) | C |
| T | Leu(1) | Ser(2) | Stop(9) | Stop(9) | A |
| T | Leu(1) | Ser(2) | Stop(9) | Try(9) | G |
| C | Leu(1) | Pro(5) | His(6) | Arg(8) | T |
| C | Leu(1) | Pro(5) | His(6) | Arg(8) | C |
| C | Leu(1) | Pro(5) | Gln(7) | Arg(8) | A |
| C | Leu(1) | Pro(5) | Gln(7) | Arg(8) | G |
| A | Ile(10) | Thr(11) | Asn(12) | Ser(2) | T |
| A | Ile(10) | Thr(11) | Asn(12) | Ser(2) | C |
| A | Met(9) | Thr(11) | Lys(13) | Arg(8) | A |
| A | Met(9) | Thr(11) | Lys(13) | Arg(8) | G |
| G | Val(14) | Ala(15) | Asp(16) | Gly(18) | T |
| G | Val(14) | Ala(15) | Asp(16) | Gly(18) | C |
| G | Val(14) | Ala(15) | Glu(17) | Gly(18) | A |
| G | Val(14) | Ala(15) | Glu(17) | Gly(18) | G |
| Function Name | Function Formula | Optimal Solution | Optimum |
|---|---|---|---|
| Ackley | [−3, 0.5] | 0 | |
| Beale | [−3, 0.5] | 0 | |
| Crossintray | [0.3459, 0.3459] | 0 | |
| Giunta | [−10, 10] | −20 | |
| Himmelblau | [3,2] | 0 | |
| Penholder | [−9.64617, 9.64617] | −0.96353 | |
| Testtubeholder | [, 0] | −10.8723 | |
| Levi13 | [1,1] | 0 |
| GA | PSO | DE | GA-TNE+DRO |
|---|---|---|---|
| G = 200 | G = 200 | G = 200 | G = 200 |
| N = 20 | N = 20 | N = 20 | N = 20 |
| Pc = 0.5 | C1 = 2, C2 = 2 | Pc = 0.5 | Pc = 0.5 |
| Pm = 0.01 | w1 = 0.9, w2 = 0.4 | Pm = 0.06 | Pm = 0.05, Pr = 0.05 |
| Function | GA | PSO | DE | GA-TNE+DRO | ||||
|---|---|---|---|---|---|---|---|---|
| 8.43 × 10−8 | 4.91 × 10−7 | 3.13 × 10−10 | 3.67 × 10−8 | 4.13 × 10−9 | 3.18 × 10−8 | 1.13 × 10−11 | 1.16 × 10−10 | |
| 3.48 × 10−7 | 6.12 × 10−6 | 5.62 × 10−9 | 4.12 × 10−8 | 7.52 × 10−10 | 7.01 × 10−9 | 7.85 × 10−11 | 8.61 × 10−10 | |
| 1.86 × 10−8 | 3.65 × 10−6 | 1.58 × 10−10 | 4.28 × 10−9 | 3.04 × 10−10 | 3.17 × 10−9 | 2.47 × 10−12 | 3.45 × 10−11 | |
| −20.0023 | −20.0238 | −19.9986 | −19.9865 | −19.9987 | −19.9928 | −20.0001 | −20.0010 | |
| 2.36 × 10−5 | 5.69 × 10−4 | 3.65 × 10−6 | 7.54 × 10−5 | 5.97 × 10−6 | 4.19 × 10−5 | 7.98 × 10−7 | 8.67 × 10−6 | |
| −0.96350 | −0.96251 | −0.96349 | −0.96332 | −0.96343 | −0.96341 | −0.96354 | −0.96350 | |
| −10.8716 | −10.8689 | −10.8718 | −10.8702 | −10.8718 | −10.8703 | −10.8721 | −10.8714 | |
| 4.35 × 10−8 | 2.36 × 10−6 | 3.68 × 10−9 | 6.39 × 10−7 | 8.26 × 10−10 | 8.69 × 10−9 | 6.31 × 10−9 | 9.23 × 10−8 | |
| Function | GA | PSO | DE | GA-TNE+DRO | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 200 | 54 | 86.8 | 200 | 51 | 65.8 | 191 | 50 | 70.4 | 180 | 9 | 15.6 | |
| 200 | 102 | 120.4 | 187 | 69 | 111.6 | 182 | 49 | 95.7 | 159 | 23 | 56.8 | |
| 198 | 118 | 142.6 | 168 | 87 | 115.6 | 174 | 37 | 61.7 | 149 | 19 | 24.6 | |
| 186 | 76 | 126.8 | 182 | 76 | 116.8 | 170 | 72 | 120.4 | 138 | 28 | 96.8 | |
| 178 | 78 | 125.6 | 186 | 42 | 87.6 | 158 | 62 | 111.3 | 127 | 26 | 75.6 | |
| 192 | 96 | 130.1 | 192 | 63 | 97.6 | 144 | 55 | 102.7 | 106 | 27 | 38.8 | |
| 187 | 112 | 168.3 | 193 | 61 | 91.3 | 162 | 28 | 75.8 | 116 | 19 | 50.2 | |
| 186 | 86 | 118.6 | 179 | 53 | 78.6 | 104 | 13 | 24.4 | 132 | 21 | 52.4 | |
| Function | T1 | GA | PSO | DE | GA-TNE+DRO | ||||
|---|---|---|---|---|---|---|---|---|---|
| T | T | T | T | ||||||
| 4.3874 × 10−3 | 9.2735 × 10−3 | 143.71 | 5.6002 × 10−3 | 35.672 | 8.1046 × 10−3 | 109.33 | 5.2750 × 10−3 | 26.105 | |
| 3.8156 × 10−3 | 8.0649 × 10−3 | 124.98 | 7.0483 × 10−3 | 95.078 | 4.8704 × 10−3 | 31.023 | 4.5875 × 10−3 | 22.702 | |
| 7.6552 × 10−3 | 1.6180 × 10−2 | 250.74 | 1.4141 × 10−2 | 190.75 | 9.7714 × 10−3 | 62.241 | 9.2038 × 10−3 | 45.547 | |
| 7.9521 × 10−3 | 1.6808 × 10−2 | 260.47 | 1.4689 × 10−2 | 198.15 | 1.0150 × 10−2 | 64.654 | 9.5607 × 10−3 | 47.313 | |
| 1.8687 × 10−3 | 3.9498 × 10−3 | 61.210 | 2.3853 × 10−3 | 15.194 | 3.4519 × 10−3 | 46.565 | 2.2467 × 10−3 | 11.119 | |
| 4.8976 × 10−3 | 1.0352 × 10−2 | 160.42 | 6.2515 × 10−3 | 39.821 | 9.0470 × 10−3 | 122.04 | 5.8884 × 10−3 | 29.14 | |
| 4.4559 × 10−3 | 9.4182 × 10−3 | 145.95 | 8.2309 × 10−3 | 111.03 | 5.6877 × 10−3 | 36.229 | 5.3573 × 10−3 | 26.512 | |
| 6.4631 × 10−3 | 1.3661 × 10−2 | 211.70 | 1.1939 × 10−2 | 161.05 | 7.7706 × 10−3 | 38.455 | 8.2498 × 10−3 | 52.549 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zang, W.; Zhang, W.; Zhang, W.; Liu, X. A Genetic Algorithm Using Triplet Nucleotide Encoding and DNA Reproduction Operations for Unconstrained Optimization Problems. Algorithms 2017, 10, 76. https://doi.org/10.3390/a10030076
Zang W, Zhang W, Zhang W, Liu X. A Genetic Algorithm Using Triplet Nucleotide Encoding and DNA Reproduction Operations for Unconstrained Optimization Problems. Algorithms. 2017; 10(3):76. https://doi.org/10.3390/a10030076
Chicago/Turabian StyleZang, Wenke, Weining Zhang, Wenqian Zhang, and Xiyu Liu. 2017. "A Genetic Algorithm Using Triplet Nucleotide Encoding and DNA Reproduction Operations for Unconstrained Optimization Problems" Algorithms 10, no. 3: 76. https://doi.org/10.3390/a10030076





