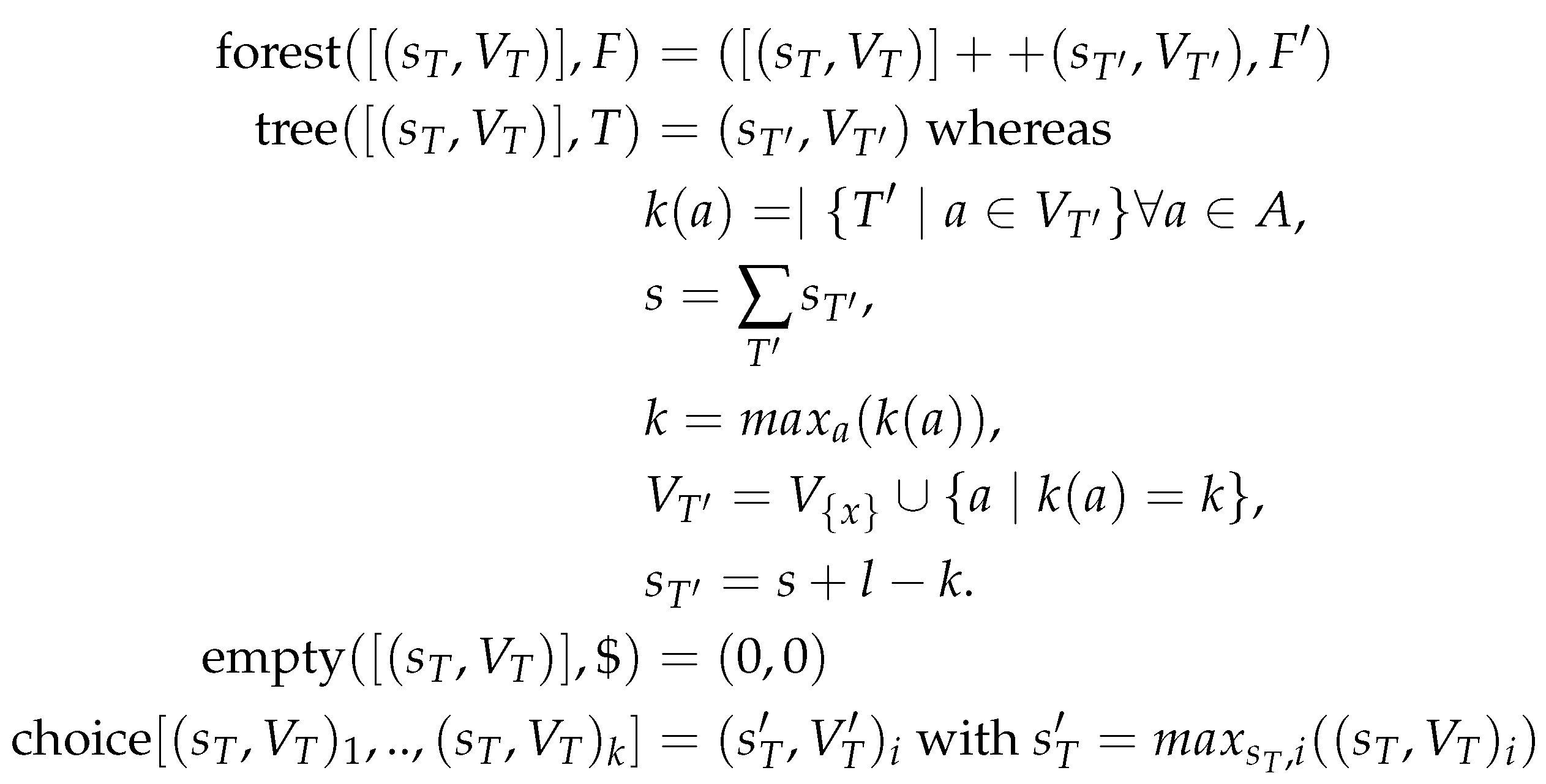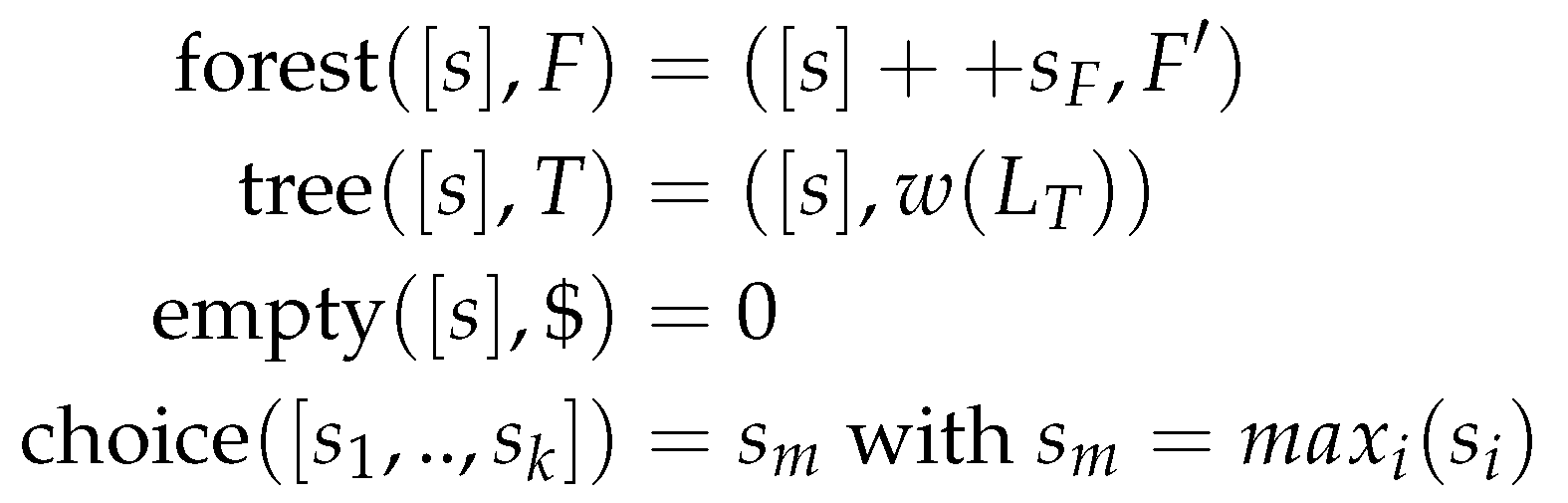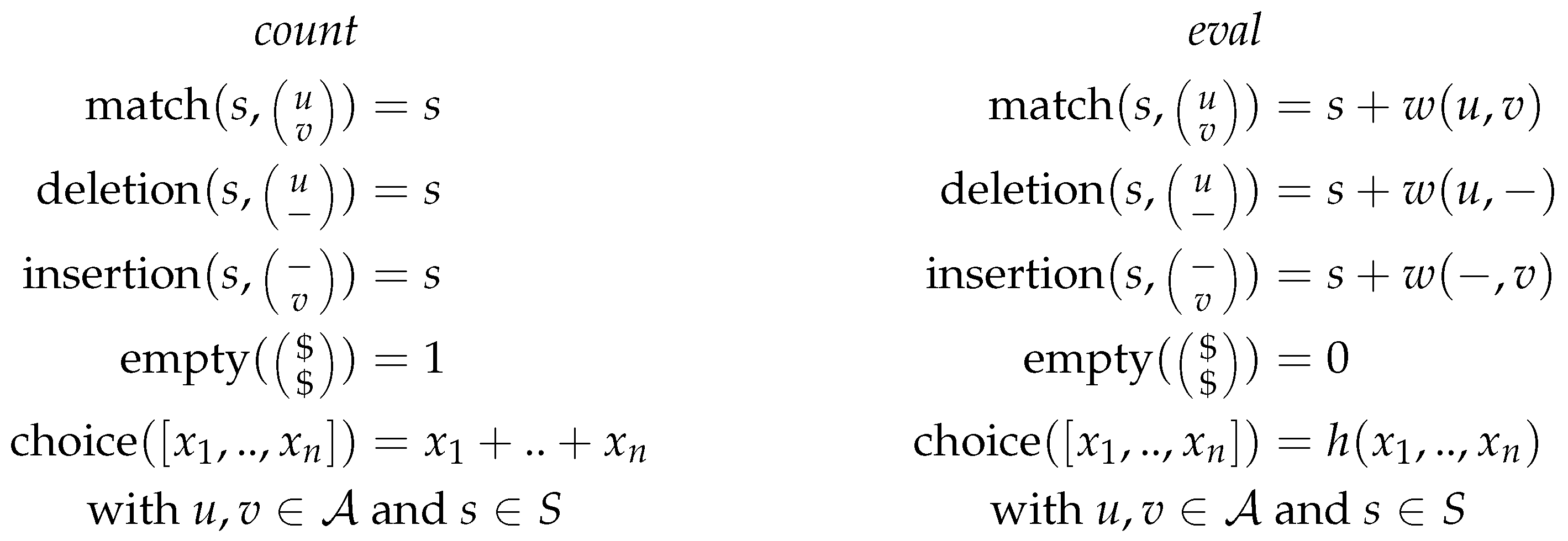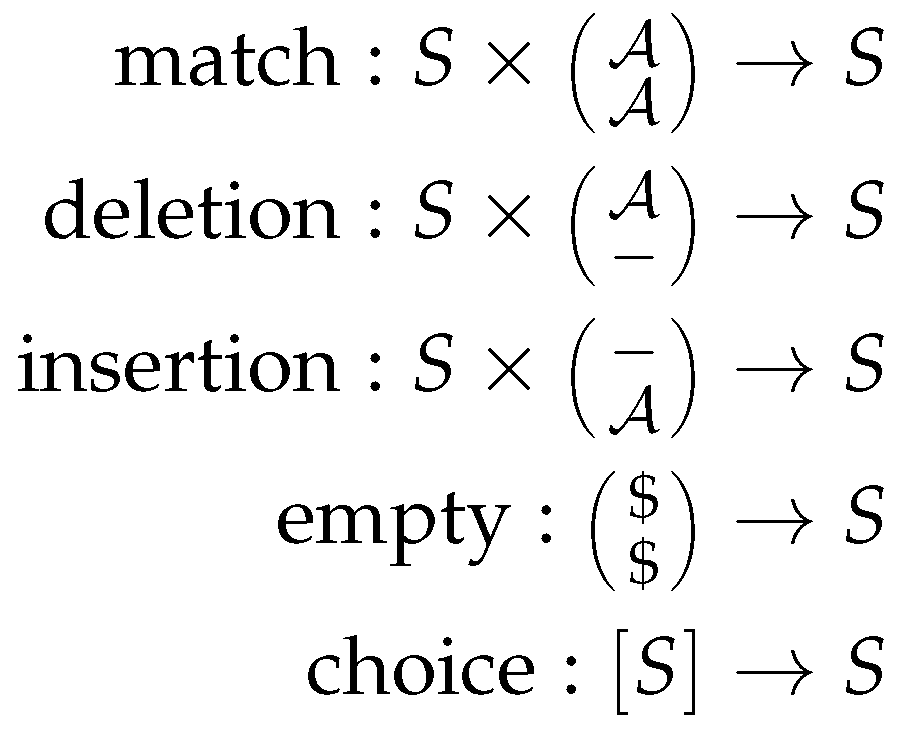1. Introduction
Dynamic programming (DP) is a general paradigm for solving combinatorial problems with a search space of exponential size in polynomial time (and space). The key idea is that the optimal solution for the overall problem can be constructed by combining the optimal solutions to subproblems (known as optimal substructure). During the recursive deconstruction of the original problem instance, the same smaller subproblems (known as overlapping subproblems) are required repeatedly and thus memoized. DP is typically applied to optimization and (weighted) enumeration problems. Not all such problems are amenable to this strategy however.
Bellman’s principle [
1] codifies the key necessary condition, the intuition of which can be expressed for optimization problems as follows: The optimal solution of any (sub)problem can be obtained as a suitable combination of optimal solutions of its subproblems.
In Bioinformatics, DP algorithms are a useful tool to solve tasks such as sequence alignment [
2] or RNA secondary structure prediction [
3,
4]. Here, the description of dynamic programming algorithms as formal grammars is common practice [
5]. Problems whose natural input structure is a tree typically have no satisfactory solution using an encoding as strings, because one “dimension” within the input is lost. As a consequence, solutions to seemingly similar string problems can be very different. A simple example is comparison of two patterns of matching parentheses:
((((((())))))) ((((((())))))--)
(((((()))))()) ((((((--)))))())
On the l.h.s., they are interpreted as strings differing by two substitutions. On the r.h.s., interpreted as trees the difference is the insertion and deletion of matching pairs in each pattern.
A simple, yet often encountered, example of a tree is the structure of a text such as a book. Books are divided into chapters, sections, subsections, and blocks of text, imposing a hierarchical structure in addition to the linear order of words. Many other formats for data exchange are based on similar structures, including
json and
xml. In bioinformatics, tree structures encode, among others, the (secondary) structure of RNA and phylogenetic trees, hence DP algorithms on trees have a long history in computational biology. Well-studied problems include the computation of parsimony and likelihood scores for given phylogenies [
6], tree alignment [
7] and tree editing [
8], reconciliation of phylogenetic trees [
9], and RNA secondary structure comparison [
10,
11]. Giegerich and Touzet [
12] developed a system of inverse coupled rewrite systems (ICOREs) which unify dynamic programming on sequences and trees. Rules written in this system bear a certain semblance to our system. We are not aware of an implementation of ICORES, however, making comparisons beyond the formalism difficult. All of these approaches use trees and/or forests as input structures. Aims and methods, however, are different. We argue here that a common high-level framework will substantially simplify the design and development of such algorithms and thus provide a very useful addition to the toolbox of someone working with tree-like data. There are implementations for most of the problems mentioned above. However, they use problem-specific choices for the recursions and memoization tables that seem to be hard to generalize. A particularly difficult challenge is the construction of DP algorithms that are not only complete but also semantically unambiguous. The latter property ensures that each configuration is considered only once, so that the DP recursion correctly counts and thus can be used to compute partition functions. A recent example is the approach of [
7] to tree alignment. In general, recognizing ambiguity for context-free grammars is undecidable [
13,
14]. There are, however, quite efficient heuristics that can be used successfully to prove that string grammars describing many problems in computational biology are unambiguous [
15]. The aim of the present contribution is to develop a general framework as a generic basis for the design and implementations of DP algorithms on trees and forests. We focus here on well-known algorithms with existing implementations as examples because in these cases the advantages of the ADP approach can be discussed directly in comparison to the literature. Since DP algorithms are usually formulated in terms of recursion equations, their structure is usually reflected by formal grammars. These are more compact than the recursions and more easily lend themselves to direct comparisons. In this way, we therefore can compare results and make sure that the algorithms are either unambiguous or work well even in the ambiguous case. Here, ambiguity does not affect the running time of the algorithms.
The most difficult issue in this context is to identify a sufficiently generic representation of the search spaces on which the individual application problems are defined. Following [
16], a natural starting point is a formal grammar
G as a rewriting system specifying a set of rules on how to generate strings or read strings of the corresponding language
. The benefit of using formal grammars compared to, say, explicit recurrences, is mostly in the high-level description of the solution space defined by the problem at hand. For formal grammars describing dynamic programming problems on strings several approaches have been developed in recent years [
16,
17,
18,
19,
20]. Formal grammars can describe tree languages just as well as string languages. This opens up the benefits of using formal languages (in abstracting away the minutia of implementation details) for this expanded input type [
21,
22].
Our contributions are twofold. First, we provide a theoretical framework for describing formal languages on trees. We give a small set of combinators—functions that formalize the recursive deconstruction of a given tree structure into smaller components—for the design of such formal languages. Second, we show that an implementation in the
ADPfusion [
18] variant of algebraic dynamic programming (ADP) [
23] yields short and elegant implementations for a number of well-known algorithms on tree structures.
With this contribution
ADPfusion now encompasses the most widely used types of inputs for dynamic programs, namely (i) strings [
17,
18], (ii) sets [
24], and (iii) trees (this work).
2. Algebraic Dynamic Programming (ADP)
Algebraic dynamic programming (ADP) [
18,
25] is designed around the idea of higher order programming and starts from the realization that a dynamic programming algorithm can be separated into four parts: (i) grammar, (ii) algebra, (iii) signature, and (iv) memoization. These four components are devised and written separately and combined to form possible solutions. One advantage of ADP is that the decoupling of the individual components makes each individual component much easier to design and implement. Another advantage is that different components can be re-used easily. This is in particular true for the grammar, which defines the search space. Hence, it needs to be specified only once. The grammar only specifies the input alphabet and the production rules on how to split the input data structure in subparts. The algebra specifies the cost function and the selection criterion, also called objective function, and thus defines formally what is considered as an “optimal” solution. Based on the objective function, the optimal solution is selected from the possible solutions. The scope of the evaluation algebra is quite broad. It can be used e.g., to count and list prioritized (sub-)optimal solutions. The signature is used as a connecting element between grammar and algebra such that they can be re-used and exchanged independently from each other as long as they fit to the signature. In order to recursively use subsolutions to the problem, they are stored using a data structure easy to save and look-up the subsolution to the current instance. This is called memoization. In this section, the four parts of ADP will be explained using string alignment as an example.
Before we delve into dynamic programming on trees and forests, we recapitulate the notations for the case of dynamic programming on strings. The problem instance is the venerable Needleman-Wunsch [
26] string alignment algorithm. The algorithm expects two strings as input. When run, it produces a score based on the number of matches, mismatches, deletions, and insertions that are needed to rewrite the first into the second input string.
One normally assigns a positive score to a match, and negative scores to mismatches, deletions, and insertions. With the separation of concerns provided by algebraic dynamic programming, the algebra that encodes this scoring becomes almost trivial, while the logic of the algorithm, or the encoding of the search space of all possible alignments in the form of a grammar, is the main problem. As such, we mostly concentrate on the grammars, delegating scoring algebras to a secondary place.
A simple version of the Needleman-Wunsch algorithm [
26] is given by:
The grammar has one 2-line symbol on the left-hand side, the non-terminal . The notion of two lines of X is indicative of the algorithm requiring two input sequences. This notion will be made more formal in the next section. Here, X is a non-terminal of the grammar, a a terminal character, $ the termination symbol and ‘−’ the “no input read”-symbol. Non-terminals such as are always regarded as one symbol, and can be replaced by any of the rules on the right-hand side that have said symbol on the left-hand side. Terminals, say , act on individual inputs, here not moving on the first input, but reading a character “into” a on the second input. In theory of automata it is customary to refer to independent inputs as “input tapes”. The operations of the grammar can be explained for each symbol individually, while grouping into columns indicates that the symbol on each line, and thereby input, operates in lockstep with the other symbols in the column. Multiple tapes thus are not processed independently, i.e., ‘actions’ are taken concurrently on all tapes. Since we consider the insertion of a gap symbol also as an ‘action’, it is not allowed to insert gap symbols on all the tapes at the same time. Hence, in each step at least one of the tapes is ‘consuming’ an input character. The algorithm is therefore guaranteed to terminate.
The grammar defines 4 different cases: (i) matching the current positions of the input strings, (ii)–(iii) inserting a gap symbol in one or the other of the input strings or the case (iv) when both input strings are empty. As the algorithm should return the best possible alignment of the input strings, each of the production rules is assigned a scoring function in the scoring algebra. The scoring function will take the current score and return an updated score including the weight for the current case (match, deletion or insertion).
Figure 1 shows two different scoring algebras for the grammar of string alignment, with
being the input alphabet, and
S the set of scores. Thus, for each grammar, we can choose a scoring algebra independently. The choice function corresponds to the objective function of the dynamic programming algorithm. As it is used to rank given (sub)solutions, it is not part of the grammar but part of the scoring algebra and the signature, as seen in
Figure 1 and
Figure 2.
The signature in ADP provides a set of function symbols that provide the glue between the grammar and the algebra. Thus, we define the type signatures for each of the scoring functions, analogously to type signatures found in functional programming.
Figure 2 shows the signature for the Needleman-Wunsch algorithm. This signature fits to both scoring algebras and the grammar. Here,
is the input alphabet, and
S the set of scores as above.
The last part needed for ADP is the memoization to store the subsolutions. For the Needleman-Wunsch algorithm we simply use a matrix of size where s and t are the lengths of the input strings to store the subsolutions.
3. DP Algorithms on Trees and Forests
Algebraic dynamic programming [
25] in its original form was designed for single-sequence problems. Alignment-style problems were solved by concatenating the two inputs and including a separator.
Originally,
ADPfusion [
18] was designed along the lines of ADP, but gained provisions to abstract over both the input type(s), and the index space with the advent of generalized algebraic dynamic programming [
17,
24,
27,
28,
29] inputs were generalized to strings and sets, as well as multiple tapes (or inputs) of those.
Trees as data structures sit “somehow between” strings and sets. They have more complex structures, in that each node not only has siblings, but also children compared to lists which only have siblings for each node in the list or string. Sets on the other hand are typically used based on the principle of having edges between all nodes, with each edge having a particular weight. This alone makes trees an interesting structure to design a formal dynamic programming environment for. Tree and forest structures have, as discussed above, important functions in (for example) bioinformatics and linguistics.
DP algorithms can be applied not only to just one input structure but also to two or more independent inputs. In the context of formal grammars describing DP algorithms we denote DP algorithms applied to one input structure as single-tape and two or more inputs as two-tape or multi-tape algorithms in analogy to the terminology used in the theory of automata in theoretical computer science. Mixing tapes, i.e., having a tree on one tape and a string on the other is possible. Due to the splitting of grammar, signature, algebra and memoization, the underlying machinery is presented with an index structure to traverse that corresponds to the decomposition rules of the data structure(s). Hence, during one iteration step it is possible that the algorithm consumes one character of the input string on one tape whereas the second tape recurses on a tree.
We now formalize the notion of an input tape on which formal grammars act. For sequences, an input tape is a data type that holds a finite number of ordered elements. The ordering is one-dimensional. It provides the ability to address each element. For all algorithms considered here, the input tape is immutable. Sequences are the focus of earlier work [
17,
18].
Input tapes on
trees hold a finite number of elements with a parent-child-sibling relationship. Each element has a single parent or is a root element. Each element has between zero and many children. The children are ordered. For each child, its left and right sibling can be defined, unless the child is the left-most or right-most child. Input tapes on
forests introduce a total order between the root elements of the trees forming the forest. These structures are discussed in this work. For sets, the tape structure is the power set and the partial order defined by subset inclusion. Set structures have been discussed earlier [
24].
A multi-tape grammar, such as the example above, extends this notion to tuples of input tapes. We will formalize the notion of multi-tape inputs later in Definition 8. We note in passing that in the case of multiple context-free grammars the one-to-one correspondence between the
i’th element of the tuple and the
i’th row of each rule does not hold for interleaved symbols. We refer to [
29] for the details.
In order to define formal grammars for DP algorithms on trees and forests, we have to define how to split a tree or a forest into its substructures. On strings, the concatenation operator is assumed implicitly to concatenate single symbols of the string. We make the operators explicit as there is more than one operator on trees and forests.
Definition 1 (trees and forests).
Let T be a rooted, ordered tree. Then, either T is empty and contains no nodes, or there is a designated node r, the root of the tree. In addition, for each non-leaf node, there is a given order of its children from left to right. Furthermore, let F be the ordered forest of trees , where the order is given by assuming a global root node , of which are the direct, ordered children. Labels are associated with both leaves and inner nodes of the tree or forest.
We now define the set of operations that will lead to dynamic programming operations on ordered trees and forests. Each operation transforms a tree or forest on the left-hand side of a rule (→) into one or two elements on the right-hand side of a rule. Rules can fail. If a rule fails, it does not produce a result on the right-hand side. In an implementation, right-hand sides typically yield sets or lists of results, where a failure of a rule naturally leads to an empty set or an empty list.
Definition 2 (empty tree or forest).
If T is empty, then the rule yields ϵ. If F is empty, then the rule yields ϵ.
Definition 3 (single root).
If T with root r contains just the root r, then yields r.
Definition 4 (root separation).
Given non-empty T, the rule yields the root r of T, as well as the ordered forest of trees. Each is uniquely defined as the k’th child of the root r of T. is isomorphic to this rule.
Definition 5 (leaf separation).
Given non-empty T, the left-most leaf that is defined as the leaf the can be reached by following the unique path from the root r of T via each left-most child until a leaf has been reached. We write for the tree obtained from T by deleting the leaf l and its incident edge. The rule breaks up T into the leaf l and the remaining tree . The rule likewise extracts the right-most leaf.
Definition 6 (forest root separation).
Let be non-empty forest and let r be the root of , the right-most tree in F. The rule separates the right-most root, that is, the resulting forest replaces in the the original forrest by the child-trees of r. The rule analogously separates the left-most root r by replacing the the leftmost tree of F by the child-trees of its root.
Definition 4 is a special case where the forest only consists of one tree.
Definition 7 (forest separation)
The rule separates the non-empty forest F into the left-most tree T, and the remaining forest . T is uniquely defined by the ordering of F. The rule likewise yields the right-most tree.
The following lemma gives that both tree and forest decomposition are well defined and always lead to a decomposition using at least one of the definitions above.
Lemma 1 (tree decomposition).
Given any (empty or non-empty) tree T, at least one of the rules Definitions 2–5 yields a decomposition of the tree, where either T is decomposed into “atomic” elements (ϵ or a single node), or into two parts, each with fewer vertices than T.
Proof. Every tree T is either empty (Definition 2), contains just one node which then is the root (Definition 3), or contains one or more nodes (Definitions 4 and 5), of which one is separated from the remainder of the structure. ☐
Lemma 2 (forest decomposition).
For every forest F the operations Definitions 2, 6 and 7 yield a decomposition.
Proof. The argument is analogously to the case of trees. The forest root separation cases given in Definition 6 remove the left- or right-most root from the forest, hence they constitute a decompostion of the structure. This might lead to a larger number of trees in the resulting forest but reduces the total number of nodes. ☐
Given those tree and forest concatenation operators, we are now able to define formal grammars to describe DP algorithms on trees and forests. In our framework, rules are either executed as a whole or fail completely. A rule failing partially is not possible.
Lemma 3 (Totality).
Given an arbitrary tree or forest, at least one of the operations specified in Definitions 2–7 and Lemmas 1 and 2 can be applied. Each of these operations reduces the number of nodes associated with each non-terminal symbol or results in a single node or an empty structure. Thus any finite input tree or forest can be decomposed by a finite number of operations.
Proof. Let F be a forest. For the case of the empty forest, we apply Definition 2. In the case of more than one tree, we apply Definition 7 which yields a single tree and another forest that are both smaller than the original forest. This is also shown in Lemma 2.
In case of Definition 7, the resulting forest structure can be empty such that the original forest only consisted of a single tree. Here, we continue with the possible tree structures.
A tree T can be empty, or consist either of a single node or more than one node. For the empty case, we apply Definition 2. In case of the tree consisting only of a single node, we apply Definition 3. If T consists of more than one node, we either apply Definition 4 or Definition 5. In case of Definition 4 the resulting structure consists of a single node (an atomic structure) and a forest which in total has a smaller number of nodes than the original tree.
In the case of Definition 5, one leaf is eliminated from the tree, thus this yields a tree that just differs from the original structure by one leaf.
For the case of algorithms working on two input structures, it is ensured that at least one of the current structures is decomposed such that the total number of nodes decreases within the next recursion step. ☐
The operators on tree and forest structures defined above specify how these data structures can be traversed. They specify a basis for the formulation of at least a large collection of DP problems using tree and forest decomposition. Different DP problems thus can be formulated (as a formal grammar) based on the same set of decomposition rules. There are many ways of designing DP algorithms on tree structures [
30,
31,
32]. In case they apply the same tree and forest decomposition rules, the index structures and operators will be same and only grammar and signature, and possible the scoring algebra, need to be edited. At present we lack a formalized theory of decompositions for arbitrary data structures hence we cannot formally prove that this set of operators is sufficient to formulate any meaningful DP problem on trees. If necessary, however, additional decomposition operators can be added to the list.
6. Tree Alignment
Our grammar for the case of tree alignment is based on the grammars formulated in [
42,
43]. We expand those grammars with explicit tree concatenation operators as shown in the previous section. This allows our algorithms to parse both trees and forests as input structures.
Furthermore, we extend the grammars with the automatic derivation of an outside algorithm [
24], described in
Section 8. The combination of the inside and outside grammar allows for easy calculation of match probabilities. Since this is automatic, any user-defined grammar can be extended in this way as well. Though we point out that this still requires careful design of the inside grammar [
7].
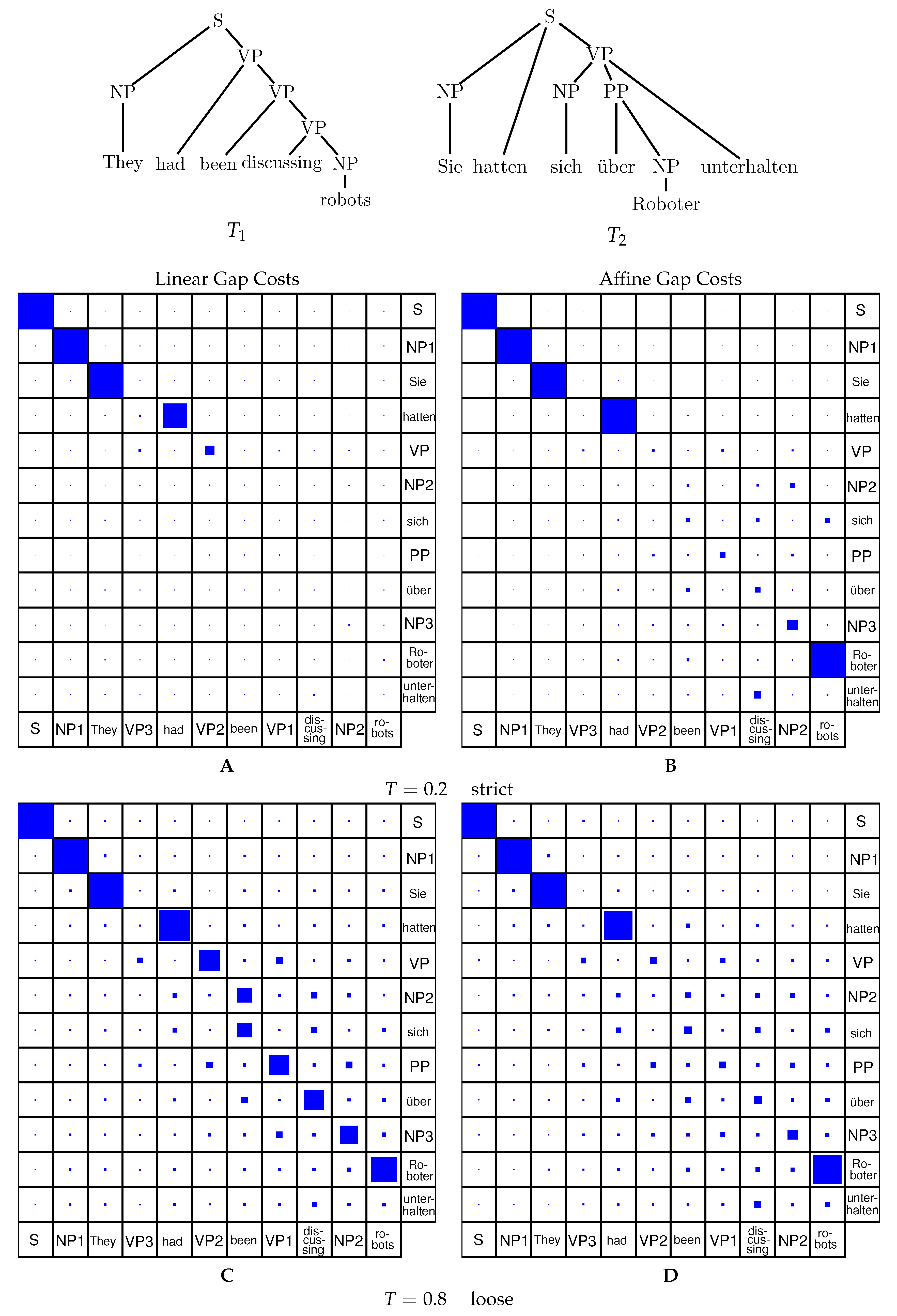
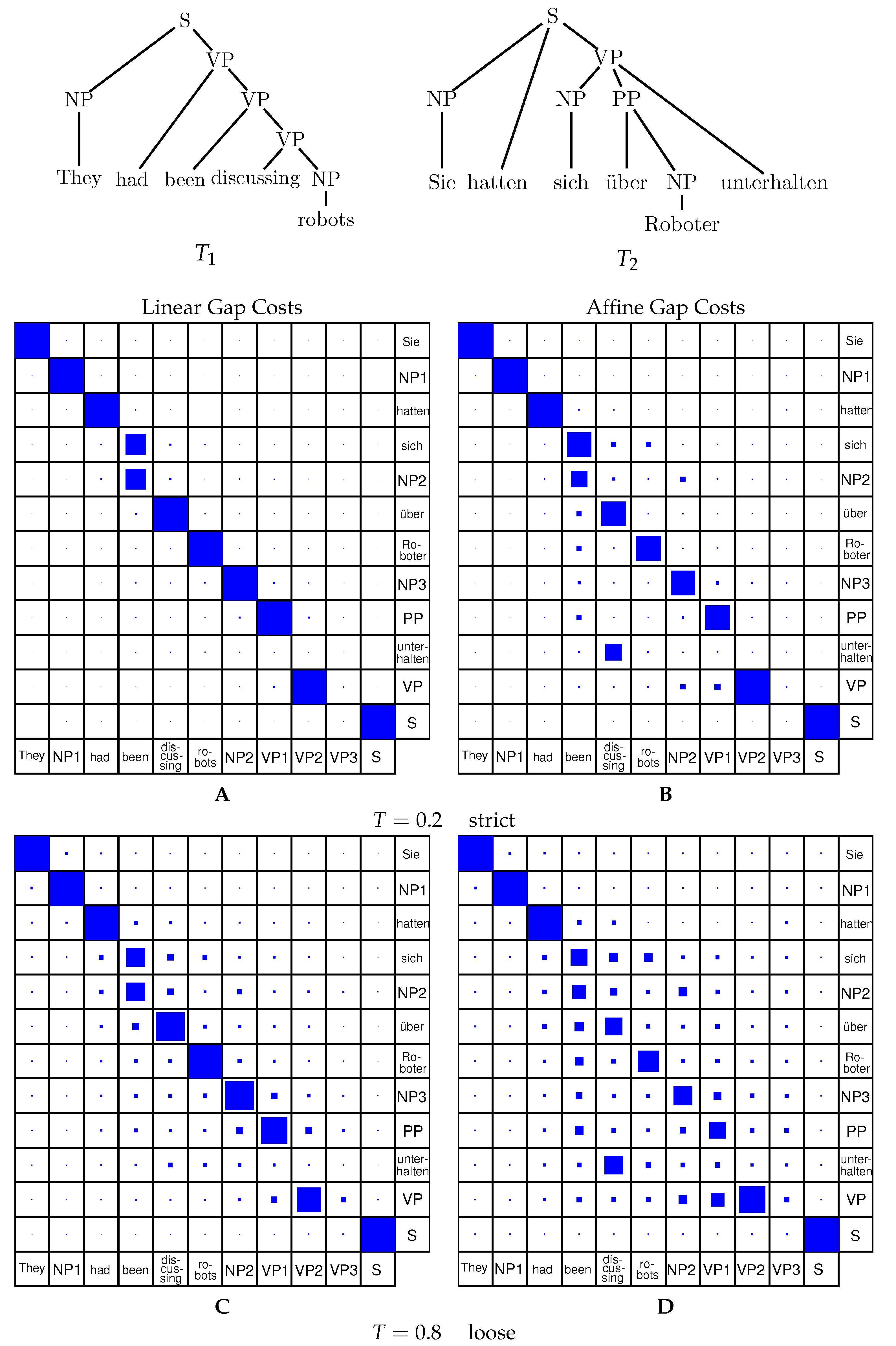
Representations of alignments (cf.
Figure 3) are typically in the form of a tree that includes nodes from both trees, where matching nodes are given as a pair and deleted nodes from both trees are inserted in a partial-order preserving fashion.
Definition 9 (Tree alignment).
Consider a forest G with vertex labels taken from with as alphabet. Then we obtain mappings and by considering only the first or the second coordinate of the labels, respectively, and by then deleting all nodes that are labeled with the gap character ‘−’, see Figure 3. G is an alignment of the two forests and if and . The cost of the alignment
G based on the original forests
and
is the sum of the costs based on the labels in
G as formulated in Equation (
5). Each label consists of a pair of labels
, whereas
corresponds to a label in the vertex set of
or a gap symbol ‘−’ and analogously for
.
Every alignment G defines unique mappings and , but the converse is not true. The minimum cost alignment is in general more costly than the minimum cost edit script.
We will need a bit of notation. Let F be an ordered forest. By we denote the subforest consisting of the first i trees, while denotes the subforest starting with the -st tree. By we denote the forest consisting of the children-trees of the root of the first tree in F. is the forest of the right sibling trees of F.
Now consider an alignment A of two forests and . Let be the root of its first tree. We have either:
. Then and ; is an alignment of and ; is an alignment of and .
. Then ; for some k, is an alignment of and , and is an alignment of with .
. Then ; for some k, is an alignment of and and is an alignment of with .
These three cases imply the following dynamic programming recursion:
with initial condition
. The formal grammar underlying this recursion is
It is worth noting that single tape projections of the form make perfect sense. Since − is a parser that always matches and returns an empty string, which in turn is the neutral element of the concatenator this formal production is equivalent to . Hence, it produces a forest F that happens to consist just of a single tree T.
As depicted in
Figure 4, fixing the left-most tree
T in a forest, there are two directions to traverse the forest: downwards towards the forests of the root’s children
and sideways towards the remaining forest
. Regarding one single forest, the subforests
and
are two disjoint entities, thus once split, they do not share any nodes in a further step of the decomposition algorithm. The grammar for tree alignment as described above is inefficient, however, because there is no explicit split between
and
in the first step. The grammar shown in Equation (
8) explicitly splits the two forests in an earlier step to avoid redundancy.
An efficient variant that makes use of a number of facts that turn this problem into the equivalent of a linear grammar on trees has been described [
44]. Trees are separated from their forests from left to right, and forests are always right-maximal. Given a local root node for the tree, and a ternary identifier ({
T,F,E}), each forest, tree, and empty forest can be uniquely identified. Trees by the local root, forests by the local root of their left-most tree, and empty forests by the leaves “above” them. The asymptotic running time and space complexity for trees with
m and
n nodes respectively is then
. This also holds for multifurcating trees as the decomposition rules do not care if the tree is a binary tree or not. The children of a node in a tree always form a forest, independent of the number of children.
If the nodes of the tree are labelled in pre-order fashion several operations on the forest can be done more efficiently. By splitting a forest into a tree and the remaining forest, we need to store the tree’s indices to know where it is located in the forest. In case of a pre-order indexing we can take the left-most tree without storing additional indices as the left border of the tree is the smallest index (which is its root) of the original forest and the right border of the tree is just the predecessor of the left-most root of the remaining forest. Thus, storing the roots’ indices in a forest will directly give us the right-most leaves of the corresponding trees.
We finally consider a variant of Equation (
7) that distinguishes the match rule
with a unique non-terminal
on the left-hand side of Equation (
8). This rule, which corresponds to the matching of the roots of two subtrees, is critical for the calculation of
match probabilities and will play a major role in
Section 8. The non-terminals
and
designate insertion and deletion states, respectively. Thus, we can formulate the formal grammar for tree alignment as follows:
This grammar explicitly splits
and
by applying
iter. Hence, these parts recurse independently from each other. As shown in
Figure 4, aligning two trees can lead to aligning the downwards forest of one tree to the forest to the right of the other tree. Given Equation (
6), the first case corresponds to our first rule in
iter and the
align rule. Thus replacing
in
iter we obtain
. Here,
will give the score for aligning two non-terminals thus
. The tree concatenation operators
and
correspond to
and
, respectively. Replacing
and
in the
deletion and
insertion rules, we get
and
. This corresponds to case 2 and 3 in Equation (
6). As depicted in
Figure 4, given a deleted root in one tree, we recurse by aligning both subforests. In case one of the subforests is empty, it will be replaced by the first tree (the first
k trees, respectively) of the remaining forest.
The following shows an example scoring algebra for the case of tree alignment with linear gap costs. No costs are added in the
iter case but scores obtained from subsolutions are added, given by
and
. Here,
U and
V stand for the non-terminal characters whereas
are terminals.
Two and Multi-Tape Tree Alignment
Definition 10 (multiple tree alignment).
Let be ordered trees following Definition 1. The partial order of nodes of each tree sorts parent nodes before their children and children are ordered from left to right. An alignment between trees is a triple of functions with defines a function matching nodes in tree to the consensus tree of the alignment:
(i) The set of matched nodes form a partial order-preserving bijection. That is, if are matched with the partial orders and hold for all nodes and pairs of trees . (ii) The set of deleted nodes form a simple surjection from a node onto a symbol (typically ‘-’) indicating no partner. (iii) For trees , (ii) holds analogously.
The problem descriptions in Definitions 9 and 10 are equivalent. Every forest can be defined as a tree with the roots of the forest as children of a trees’ root. Given Definition 9, the mappings and can therefore be applied to a tree as well.
Given Definition 10 (i), the partial order is also preserved in case of forests, as the roots of the forests are partially ordered, too. Definition 10 (ii) and (iii) define the order of the deleted nodes that is preserved within the tree. This is also true for forests, and obtained by the mappings and defined in Definition 9.
A multiple alignment of forest structures can be constructed given the Definitions 2–7 and Lemmas 1 and 2 We now show that, given these definitions, the alignment is well-defined and can always be constructed independent of the input structures. We do not show here that the resulting alignment is optimal in some sense, as this depends on the optimization function (or algebra) used.
Let be an input of k forests. Successive, independent application of the operations specified in Definitions 2–7 and Lemmas 1 and 2 to individual input trees yields the multi-tape version of the empty string , i.e., the input is reduced to empty structures. Not every order of application necessarily yields an alignment.
Just as in the case of sequence alignments [
17] the allowed set of operations needs to be defined for multi-tape input. This amounts in particular to specify which operations act concurrently on multiple tapes. The above-given definitions can be subdivided into two sets. One set of operations takes a forest
F and splits off the left- or right-most tree (
or
), where both
and
T are allowed to be empty. In case of
T being empty then it holds that
. If
is empty, then this yields a decomposition of
F only if
F consists of just a single tree. The other set of operations removes atomic elements from a tree and yields a tree or forest, depending on the rule used.
As in [
17], all operations have to operate in “type lockstep” on all inputs simultaneously. That is, either all inputs separate off the left- or right-most tree and yield the trees
and remaining forests
, or all operations remove a terminal or atomic element, yielding the terminal elements
and remaining structure
.
Furthermore, an operation ‘−’ is introduced. This operation does not further decompose T but provides a terminal symbol ‘−’ as element . This is analogous to the in/del case in string alignments.
Theorem 1 (Alignment).
Every input of forests can be deconstructed into by successive application of the steps outlined above.
Proof of Theorem 1. At least one of the decompositions defined above for forests (Definitions 2, 6 and 7) or trees (Definitions 2–5) can always be applied for each input. Due to lockstep handling, all inputs simultaneously separate into a tree and remaining forest (yielding two structures), or apply a rule yielding a terminal element. As such, either all structures are already empty () or at least one structure can be further decomposed yielding a smaller structure.
11. Implementation
Now that all required theory is in place, we turn toward a prototypical implementation. During the exposition we make several simplifications that are not present in the actual implementation. We also suppressed all references to monadic parametrization. The entire framework works for any monad, giving a type class class (Monad m) => MkStream m x i below. While this is powerful in practice, it is not required in the exposition. As such, the MkStream type class will be presented simply as MkStream x i.
Ordered forests are implemented as a set of vectors instead of using the more natural recursive structure in order to achieve asymptotically better access.
data TreeOrder = Pre | Post
data Forest (p :: TreeOrder) a where
Forest
{ label :: Vector a
, parent :: Vector Int
, children :: Vector (UVector Int)
, lsib :: Vector Int
, rsib :: Vector Int
, roots :: Vector Int
} -> Forest p a
Each node has a unique index in the range
, and is associated with a label of type a, a parent, a vector of children, and its left and right sibling. In addition, we store a vector of roots for our forest. The Vector data type is assumed to have an index operator (!) with constant time random access. Forests are typed as either being in pre-order or post-order sorting of the nodes. The linear grammars presented beforehand make direct use of the given ordering and we can easily guarantee that only inputs of the correct type are being handled. The underlying machinery for tree alignments is more complex and our choice for an example implementation in this paper. The implementation for tree edit grammars is available in the sources.
Tree Alignment. The tree alignment grammar is a right-linear tree grammar and admits a running time and space optimization to linear complexity for each tape. The grammar itself has been presented in
Section 6. Here, we discriminate between three types of sub-structures within a forest via
data TF = T | F | E
which allows a forest to be tagged as empty (
E) at any node
k, be a single tree
T with local root
k, or be the subforest
F starting with the left-most tree rooted at
k and be right-maximal. We can thus uniquely identify sub-structures with the index structure
data TreeIxR a t = TreeIxR (Forest Pre a) Int TF
for right-linear tree indices, including the
Forest structure introduced above. The phantom type
t allows us to tag the index structure as being used in an inside (
I) or outside (
O) context.
Since the
Forest structure is static and not modified by each parsing symbol, it is beneficial to separate out the static parts.
ADPfusion provides a data family for running indices
RunIx i to this end. With a new structure,
data instance RunIx (TreeIxR (Forest Pre a) I) = Ix Int TF
we capture the variable parts for inside grammars. Not only do we not have to rely on the compiler to lift the static
Forest Pre a out of the loops but we also can implement a variant for outside indices with
data instance RunIx (TreeIxR (Forest Pre a) O)
= Ix Int TF Int TF
Outside index structures need to carry around a pair of indices. One (Int,TF) pair captures index movement for inside tables and terminal symbols in the outside grammar, the other pair captures index movement for outside tables.
Regular shape-polymorphic arrays in Haskell [
53] provide inductive tuples. An inductive tuple
(Z:.Int:.Int) is isomorphic to an index in
, with
Z representing dimension 0, and
(:.) constructs inductive tuples. A type class
Shape provides, among others, a function
toIndex of the index type to a linear index in
. We make use of an analogous type class (whose full definition we elude) that requires us to implement both a
linearIndex and a
size function. The linear index with largest (
) and current (
) index in the forest
linearIndex (TreeIxR _ u v) (TreeIxR _ k t)
= (fromEnum v + 1) * k + fromEnum t
and size function
size (TreeIxR _ u v) = (fromEnum v + 1) * (u+1)
together allow us to define inductive tuples with our specialized index structures as well.
linearIndex and
size are defined for the tuple constructor
(a:.b) and combine both functions for
a and
b in such a way as to allow us to write grammars for any fixed dimension.
Parsing with Terminal Symbols. In a production rule of the form
, terminal symbols, denoted t perform the actual parsing, while non-terminals (L, R) provide recursion points (and memoization of intermediate results).
For tree grammars, three terminal symbols are required. The $-parser is successful only on empty substructures, the deletion symbol (−) performs no parse on the given tape, while only the Node parser performs actual work. The first two parsers can be constructed easily, given a node parser. Hence, we give only its construction here.
First, we need a data constructor
data Node r x where
Node (Vector x -> Int -> r) (Vector x) -> Node r x
that allows us to define a function from an input vector with elements of type
x to parses of type
r, and the actual input vector of type
x. While quite often
, this generalization allows to provide additional context for the element at a given index if necessary. We can now bind the labels for all nodes of given input forest
f to a node terminal:
let node frst = Node (!) (label frst)
where
(!) is the index operator into the label vector.
Construction of a Parser for a Terminal Symbol. Before we can construct a parser for a rule such as
, we need a bit of machinery in place. First, we decorate the rule with an evaluation function (
f) from the interface (see
Section 2), thus turning the rule into
. The left-hand side will not play a role in the construction of the parser as it will only be bound to the result of the parse.
For the right-hand side we need to be able to create a stream of parses. Streams enable us to use the powerful stream-fusion framework [
54]. We will denote streams with angled brackets
<x> akin to the usual list notation of
[x] in Haskell. The elements
x of our parse streams are complicated. The element type class
class Element x i where
data Elm x i :: *
type Arg x :: *
getArg :: Elm x i -> Arg x
getIdx :: Elm x i -> RunIx i
allows us to capture the important structure for each symbol in a parse. For the above terminal symbol
Node we have
instance (Element ls i) => Element (ls , Node r x) i where
data Elm (ls,Node r x) i = ENd r (RunIx i) (Elm ls i)
type Arg (ls,Node r x) = (Arg ls , r)
getArg (ENd x _ ls) = (getArg ls , r)
getIdx (ENd _ i _ ) = i
Each Elem instance is defined with variable left partial parses (ls) and variable index (i) as all index-specific information is encoded in RunIx instances.
Given a partial parse of all elements to the left of the node symbol in
ls, we extend the
Elm structure for
ls inductively with the structure for a
Node. The Haskell compiler is able to take a stream of
Elm structures, i.e.,
<Elm x> and erase all type constructors during optimization. In particular, the data family constructors like
ENd are erased, as well as the running index constructors, say
Ix Int TF. This means that
Elm (ls, Node r x) (RunIx (TreeIxR (Forest Pre a) t))
has a runtime representation in a stream fusion stream that is isomorphic to
(r,(Int,TF),ls) and can be further unboxed, leading to efficient, tight loops. In case forest labels are unboxable
r will be unboxed as well. The recursive content in
ls receives the same treatment, leading to total erasure of all constructors.
The
MkStream type class does the actual work of turning a right-hand side into a stream of elements. Its definition is simple enough:
class MkStream x i where
mkStream :: x -> StaticVar -> i -> <Elm x i>
Streams are to be generated for symbols x and some index structure i. Depending on the position of x in a production rule, it might be considered static – having no right neighbor, or variable. In
we have D in variable and R in static position. data StaticVar = Static | Var takes care of this below.
We distinguish two types of elements x for which we need to construct streams. Non-terminals always produce a single value for an index i, independent of the dimensionality of i. Terminal symbols need to deconstruct the dimension of i and produce independent values (or parses) for each dimension or input tape.
Multi-tape Terminal Symbols. For terminal symbols, we introduce yet another inductive tuple,
(:|) with zero-dimensional symbol
M. We can now construct a terminal that parses a node on each tape via
M :| Node (!) i1 :| Node (!) i2. The corresponding
MkStream instance hands off work to another type class
TermStream for tape-wise handling of each terminal:
instance (TermStream (ts:|t) => MkStream (ls , ts:|t) i
where mkStream (ls , ts:|t) c i
= fromTermStream
. termStream (ts:|t) c i
. prepareTermStream
$ mkStream ls i
The required packing and unpacking is performed by fromTermStream and prepareTermStream, and the actual handling is done via TermStream with the help of a type family to capture each parse result.
type family TArg x :: *
type instance TArg M = Z
type instance TArg (ts:|t) = TermArg ts :. TermArg t
class TermStream t s i where
termStream :: t -> StaticVar -> i -> <(s,Z,Z)> -> <(s,i,(TArg t))>
Now, we can actually implement the functionality for a stream of
Node parses on any given tape for an inside (
I) tree grammar:
instance TermStream (ts :| Node r x) s (is:.TreeIxR a I)
where
termStream (ts:|Node f xs) (_both) (is:.TreeIxR frst i tfe)
= map (\(s, ii, ee) ->
let Ix l _ = getIndex s (Proxy :: Proxy (RunIx (is:.TreeIxR a I)))
l’ = l+1
ltf’ = if null (children frst ! l) then E else F
in (s, (ii:.Ix l’ ltf’) (ee:.f xs l)))
. termStream ts is
. staticCheck (tfe == T)
We are given the current partial multi-tape terminal state (s,ii,ee) from the left symbol (s), the partial tape index for dimensions 0 to the current dimension
(ii), and parses from dimension 0 up to
(ee).
Then we extract the node index l via getIndex. getIndex makes use of the inductive type-level structure of the index (is:.TreeIxR a I) to extract exactly the right index for the current tape. Since all computations inside getIndex are done on the type level, we have two benefits: (i) it is incredibly hard to confuse two different tapes because we never match explicitly on the structure is of the inductive index structure for the previous
dimensions. (ii) the recursive traversal of the index structure (is:.TreeIxR a I) is a type-level operation. The runtime representation is just a set of unboxed parameters of the loop functions. This means that getIndex is a runtime “no-op” since there is no structure to traverse.
For a forest in pre-order, the first child of the current node l is just
. In case of a node without children, we set this to be empty (E), and otherwise we have a forest starting at
. We then extend the partial index ii to include the index for this tape, and analogously extend the partial parse ee with this parse.
Non-Terminals. Streams are generated differently for non-terminals. A non-terminal captures the parsing state for a complete (multi-dimensional) index, not for individual tapes. The
ITbl i x data type captures memoizing behaviour for indices of type
i, and memoized elements of type
x. One may consider this data type to behave essentially like a memoizing function of type
. We can capture this behaviour thus:
instance (Elem ls i) => Elem (ls, ITbl i x) i where
data Elm (ls, ITbl i x) i = EIt x (RunIx i) (Elm ls i)
type Arg (ls, ITbl i x) = (Arg ls, r)
getArg (EIt x _ ls) = (getArg ls, r)
getIdx (EIt _ i _ ) = i
instance MkStream (ls, ITbl (is:.i) x) (is:.i) where
mkStream (ls, ITbl t f) ix
= map ((s,tt,ii) -> ElmITbl (t!tt) ii s)
. addIndexDense ix
$ mkStream ls ix
Again, we require the use of an additional type class capturing addIndexDense. In this case, this makes it possible to provide n different ways on how to memoize (via ITbl for dense structures, IRec if no memoization is needed, etc.) with m different index types using just
instances instead of
.
class AddIndexDense i where
addIndexDense :: i -> <s Z> -> <s i>
Indexing into non-terminals can be quite involved however, and tree structures are no exception. A production rule
splits a forest into the tree-like element
D that is explored further downwards from its local root, and the remaining right forest
R. Both
D and
R can be in various states. These states are
data TFsize s = EpsFull TF s | FullEps s | OneRem s
| OneEps s | Finis
In the implementation below, the optimizer makes use of constructor specialization [
55] to erase the
TFsize constructors, while they allow us to provide certain guarantees of having captured all possible ways on how to partition a forest.
Excluding Finis, which denotes that no more parses are possible, we consider four major split operations:
-
EpsFull denotes an empty tree D meaning that the “left-most” element to be aligned to another subforest will actually be empty. This will later on induce a deletion on the current tape, if an empty D needs to be aligned to a non-empty structure.
-
FullEps will assign the complete remaining subforest to D, which will (obviously) not be a tree but a forest. While no tree (on another tape) can be aligned to a forest, the subforest below a tree on another tape can be aligned to this forest.
-
OneRem splits off the left-most tree to be bound to D, with the remaining siblings (the tail) being bound to R.
-
Finally, OneEps takes care of single trees masquerading as forests during a sequence of deletions on another tape.
All the logic of fixing the indices happens in the variable case for the D non-terminal. The static case for R just needs to extract the index and disallow further non-terminals from accessing any subforest as all subforests are right-maximal.
instance AddIndexDense (is:.TreeIxR a I) where
addIndexDense (vs:.Static) (is:.TreeIxR _ _ _)
= map go . addIndexDense vs is where
go (s,tt,ii) =
let t = getIndex s (Proxy :: Proxy (RunIx (is:.TreeIxR a)))
in (s, tt:.t, ii:.Ix maxBound E)
-- continued below
In the variable case, we need to take care of all possible variations. The use of
flatten to implement “nested loops” in Haskell follows in the same way as in [
18].
Done and
Yield are stream fusion step constructors [
54] that are explicit only in
flatten.
-- continued
addIndexDense (vs:.Variable) (is:.TreeIxR f j tj)
= flatten mk step . addIndexDense vs is where
mk = return . EpsFull jj
-- forests
step (EpsFull E (s,t,i))
= Yield (s, t:.TreeIxR f j E, i:.Ix j E) Finis
step (EpsFull F (s,t,i))
= let Ix k _ = getIndex s (Proxy :: Proxy (RunIx (is:.TreeIxR a)))
in Yield (s, t:.TreeIxR f k E, i:.Ix k F) (FullEps (s,t,i))
step (FullEps (s,t,i))
= let Ix k _ = getIndex s (Proxy :: Proxy (RunIx (is:.TreeIxR a)))
u = maxBound
in Yield (s, t:.TreeIxR f k F, i:.Ix u E) (OneRem (s,t,i))
step (OneRem (s,t,i))
= let Ix k _ = getIndex (Proxy :: Proxy (RunIx (is:.TreeIxR a)))
l = rightSibling f k
in Yield (s, t:.TreeIxR f k T, i:.Ix l F) Finis
-- trees
step (EpsFull T (s,t,i))
= let Ix k _ = getIndex (Proxy :: Proxy (RunIx (is:.TreeIxR a)))
in Yield (s,tt:.TreeIxR f k E,ii:.Ix k T) (OneEps (s,t,i))
step (OneEps (s,t,i))
= let Ix k _ = getIndex (Proxy :: Proxy (RunIx (is:.TreeIxR a)))
in Yield (s,tt:.TreeIxR f k T,ii:.Ix k E) Finis
rightSibling :: Forest -> Int -> Int
rightSibling f k = rsib f ! k
This concludes the presentation of the machinery necessary to extend
ADPfusion to parse forest structures. The full implementation includes an extension to Outside grammars as well, in order to implement the algorithms as described in
Section 8. We will not describe the full implementation for outside-style algorithms here as they require some effort. The source library has extensive annotations in the comments that describe all cases that need to be considered.
The entire low-level implementation given above can remain transparent to a user who just wants to implement grammars on tree and forest structures since the implementation works seamlessly together with the embedded domain-specific languages
ADPfusion [
18] and its abstraction that uses a quasi-quotation [
56] mechanism to hide most of this more intermediate-level library.