1. Introduction
Computer aided diagnosis (CADx) can be defined as a diagnosis that is made by a radiologist who uses the output from a computerized analysis of medical images as a “second opinion” in both detecting lesions and making diagnostic decisions [
1]. One aim of the typical CADx system is to extract and analyze the characteristics of benign and malignant lesions in an objective manner to aid the radiologist. Here, the “diagnostic” decision relates to treatment response and early classification of drug responders versus non-responders, and we name our proposed system as computer-aided therapeutic response (CADrx) system.
Glioblastoma multiforme (GBM) is the most aggressive and lethal primary brain tumor in human. Anti-angiogenesis drugs are increasingly being explored in clinical trials as therapeutic options. In a phase II in vivo clinical trial, the conventional way to assess treatment response is the tumor size change after chemotherapy or radiotherapy based on Macdonald criteria and evaluated on T1-weighted contrast enhanced (T1wCE) MR images. However, efficacy can only be evaluated at least 8–10 weeks after treatment.
Diffusion weighted magnetic resonance imaging (DW-MRI) has the potential to work as a surrogate biomarker to reveal changes in the tumor microenvironment that precede morphologic tumor changes [
2]. DW-MRI depends on the microscopic mobility of water. This mobility, classically called Brownian motion, is due to thermal agitation and is highly influenced by the cellular environment of water. Because water diffusion is strongly affected by molecular viscosity and membrane permeability between intra- and extracellular compartments, DW-MRI can be used to characterize highly cellular regions of tumors versus acellular regions. Treatment response detection can be manifested as a change in tumor cellularity, which may precede tumor size changes. Thus, findings on DW-MRI could be an early sign of biologic changes. [
3]
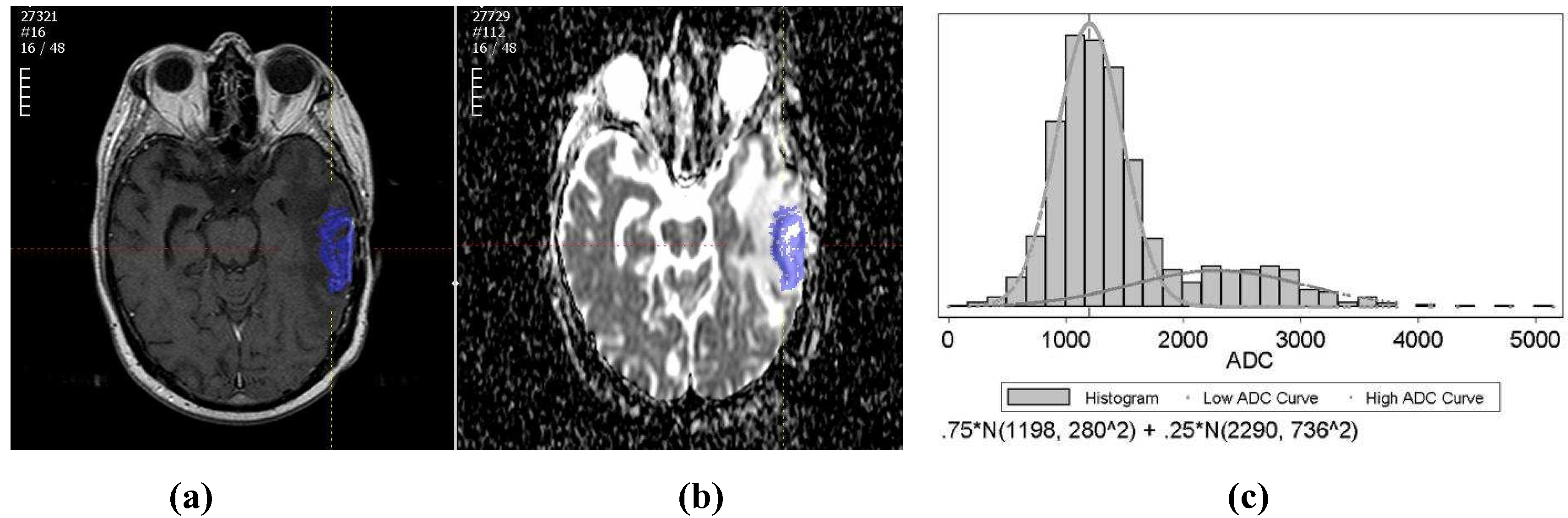
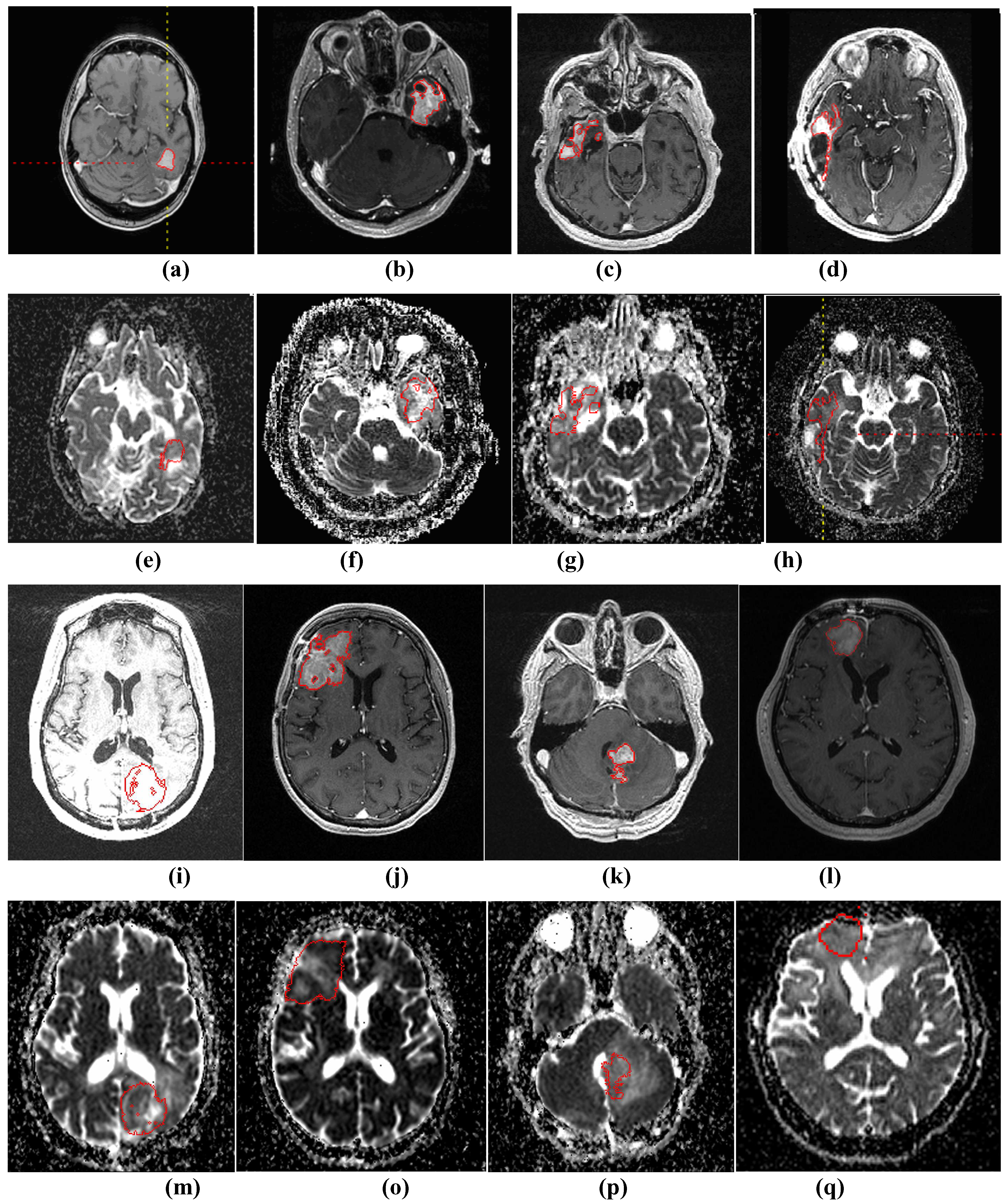
The purpose of this study is to use apparent diffusion coefficient (ADC), derived from DW-MR images, for early prediction of the tumor volume change on a later scan. There are two main parts to this computer-aided treatment response evaluation system. First, a semi-automated segmentation algorithm is applied to segment the GBM brain tumors on T1wCE images. Then, the tumor ROI is mapped onto derived ADC maps and the histogram of tumor ADC values will be extracted for automatic treatment response prediction.
Computer-aided detection and segmentation of GBM brain tumors is a challenging problem and in
Table 1 we present a concise review of the prior art in automatic tumor segmentation. Fuzzy clustering and knowledge-based analysis are popular methods explored by the early pioneers [
4,
5,
6]. Voxel-based classification method using statistical pattern classification techniques are explored by others [
7,
8,
9,
10,
11,
12,
13,
14,
15]. Most of the studies above use multiple MRI sequences (T1w, T2w, proton density weighted, and Flair) for the automatic tumor and edema detection and segmentation. Liu
et al. [
16] developed an interactive system adapting the fuzzy connectedness using multiple MRI sequences. Dube
et al. [
17,
18] used texture features and segmentation by the weighted aggregation (SWA) method for the GBM tumor segmentation on T1wCE images which is similar to part of our study. In our study, we developed semi-automated method to segment tumors on T1wCE images; in addition, we mapped the tumor contours onto ADC maps.
Table 1.
Summary of related methods in brain tumor segmentation. The type abbreviations are NC: Nasopharyngeal carcinoma; MNG: Meningiomas; MG - malignant gliomas; MS – multiple sclerosis.
Table 1.
Summary of related methods in brain tumor segmentation. The type abbreviations are NC: Nasopharyngeal carcinoma; MNG: Meningiomas; MG - malignant gliomas; MS – multiple sclerosis.
| Authors | Technique | Type | Image sequences | # of tumors |
|---|
| Liu et al. [16] | Semi-automated fuzzy clustering | GBM | T1w, T1w+c, Flair | 5 |
| Philips et al. [4] | Fuzzy clustering | GBM | PD,T2w, T1w+c | 1 |
| Clark et al. [5] | Fuzzy clustering and knowledge-based analysis | GBM | PD,T2w, T1w+c | 7 |
| Fletcher-Heath et al. [6] | Fuzzy clustering and knowledge-based analysis | Brain tumor | PD,T2w, T1w with no contrast | 4 |
| Prastawa et al. [7] | Learn distribution of normal tissues/outlier detection as tumors | Brain tumors | T2w, T1w (with or without contrast) | 3 |
| Kaus et al. [8] | Adaptive template -moderate technique with atlas prior | LGG/MG | T1w+c, sagittal view | 20 |
| Lee et al. [11] | Conditional random field and support vector machine | Brain tumors | T1w, T1w+c, T2w | 7 |
| Ho et al. [9] | 3D level set | GBM | T1w+c, T1w, T2w | 3 |
| Vinitski et al. [10] | k-nearest neighbor | MS and MG | PD, T2w, T1w, magnetization transfer | 9 |
| Zhu & Yan et al. [12] | Hopfield neural network | Brain tumors | NA | 2 |
| Zhang et al. [13] | Support vector machine | NC | T1w, T1w+c | 9 |
| Corso et al. [15] | SWA-segmentation by weighted aggregation. | GBM | T2w, T1w, T1w+c, Flair | 20 |
| Dube et al. [17] | SWA with texture features | GBM | T1w+c | NA |
| Nie et al. [14] | Spatial accuracy-weighted hidden Markov field and EM to solve the problem of high and low resolution problem | Gliomas | High:T1w, T1w+c Low:T2w, Flair | 15 |
Computer-aided diagnosis (CADx) in GBM brain tumor is an active research area, and many promising MR methods have been developed for detecting and characterizing cancer, its treatments and adverse effects, e.g. T1-weighted MR, T2-weighted MR, MR spectroscopy, perfusion-weighted MR, and diffusion-weighted MR. In our study, we focused on T1-weighted and DW-MRI. Tumor size change on T1w images is the only imaging biomarker that is accepted by the FDA as a surrogate endpoint of clinical outcome after chemotherapy and radiotherapy for phase III trials [
19]. Diffusion MRI has been explored as early detection of human GBM brain tumor treatment response early therapeutic responses before the tumor size changes.
Table 2 presents a review of the recent studies that used DWI for GBM early prediction of treatment response. Ross et al reported ADC value increase significantly in effective therapeutic intervention in pre-clinical studies and presented two patients to support this hypothesis in a preliminary clinical study [
2,
20]. Mardor
et al. [
21] applied both low and high b-value and used mean ADC and diffusion index for treatment response evaluation. Moffat et al calculated voxel-by-voxel tumor ADC value changes over time and displayed it as a functional diffusion map for correlation with clinical response [
22,
23]. They reported that the number of voxels with increased ADC is related to treatment efficacy. Our previous work [
24] showed promising results for using ADC histogram analysis, and we explored a more sophisticated classifier and designed experiments to show the advantages of the two-component histogram modeling.
Table 2.
Summary of related methods in GBM tumor treatment response using DWI.
Table 2.
Summary of related methods in GBM tumor treatment response using DWI.
| Authors | # Of Patients |
|---|
| Chenevert et al. [20] | 2 |
| Ross et al. [2] | 2 |
| Mardor et al. [21] | 10 |
| Moffat et al. [22] | 20 |
| Hamstra et al. [23] | 34 |
Machine learning and statistical pattern recognition have great contributions to the biomedical community because they can improve the sensitivity and/or specificity of detection and diagnosis of disease, while at the same time increasing objectivity of the decision-making process [
26]. The need for machine learning is perhaps greater than ever given the dramatic increase in medical data being collected, new detection, and diagnostic modalities being developed as well as the complexity of the data types and importance of multimodal analysis. In all of these cases, machine learning can provide new tools for interpreting the high-dimensional and complex datasets with which the clinician is confronted [
26]. In our study, we explored three different classification methods: AdaBoost, random forest, and support vector machine.
The AdaBoost algorithm, introduced by Freund and Schapire [
27], is an iterative algorithm that can boost weak classifiers into a strong classifier and improve the final accuracy. In each iteration, a feature is working as a weak classifier and the best feature is selected to minimize the average training error. Afterwards, the weights on training samples are redistributed in such a way that the weight of accurately classified samples will be reduced while the weight of ill classified samples is raised. Therefore, AdaBoost focuses on the most “difficult” ones [
28]. The final classifier aggregates the selected weak classifier from each iteration, and the weight for each weak classifier depends on its error rate. However, AdaBoost can be sensitive to noise and may introduce the overfitting problem.
Random forests (RF) is a classifier that combines many decision trees [
29]. Each tree depends on values of a random vector sampled independently and with equal distribution. Each tree casts a unit vote for the most popular case at input, and random forests outputs the class that is the mode of the classes output by individual trees. Breiman suggests the generalization error for forests converges to a limit as the number of trees in the forest becomes large [
30]. The error of a forest of tree classifiers depends on the strength of the individual trees in the forest and the correlation between them. Using a random selection of features to split each node yields error rates that compare favorably to Adaboost but are more robust with respect to noise.
Support vector machines (SVMs) are a set of related supervised learning methods used for classification and regression [
31,
32]. Viewing input data as two sets of vectors in an n-dimensional space, an SVM will construct a separating hyperplane in that space, one which maximizes the margin between the two data sets. To calculate the margin, two parallel hyperplanes are constructed, one on each side of the separating hyperplane, which are "pushed up against" the two data sets. Intuitively, a good separation is achieved by the hyperplane that has the largest distance to the neighboring data points of both classes, since in general the larger the margin the lower the generalization error of the classifier [
33]. SVM have been reported to work well for pharmaceutical data analysis [
34].
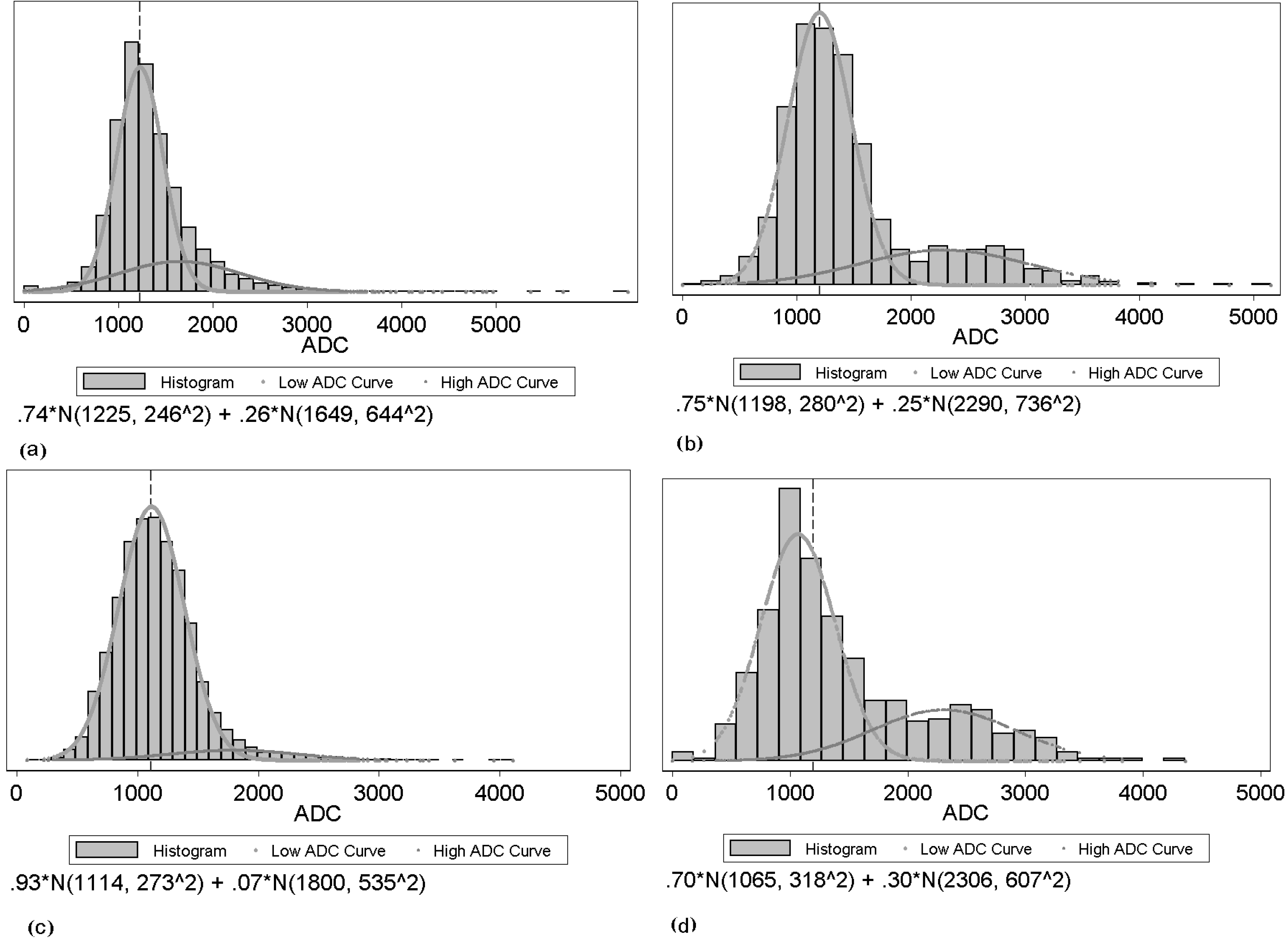
There are two main challenges in this work. One challenge is the two competing effects in ADC changes after treatment. In general, water movement inside cells is more restricted than outside. Thus, increased cell density tends to lower ADC values, whereas increased edema (more interstitial water) results in higher ADC values. Therefore, theoretically, ADC values in treated brain tumors could not only increase due to the cell kill (and thus reduced cell density), but also decrease due to inhibition of edema. None of the listed studies above have specified the separate effects. Thus, we applied a two-component model to fit the tumor ADC histogram [
25]. The other challenge is that it is difficult to directly identify GBM brain tumors on ADC maps. We developed a semi-automated framework to achieve that goal.
There are several contributions in this work. First, we developed a computer-aided method to semi-automatically identify tumors on ADC maps. Second, we explored the changes of different statistical features of the whole tumor ADC histogram. Moreover, we applied a two-component Gaussian mixture modeling to fit the tumor ADC histogram to overcome the two competing effects. Next, we used earth mover’s distance (EMD) to directly measure the distance between the pre- and post-treatment tumor ADC histograms. Finally, we introduced machine learning technique to do feature selection and classification to classify responders and non-responders.
This paper is organized as follows:
Section 2 describes the image acquisition and patient group,
Section 3 describes the semi-automated identification of GBM tumors on ADC maps, and
Section 4 describes the histogram feature extraction and classification. The Result Section reports the performance of the tumor mapping on ADC maps, and the results of our comparative study for three different classifiers. The final section offers a discussion of the experimental results as well as the future work.
6. Discussion
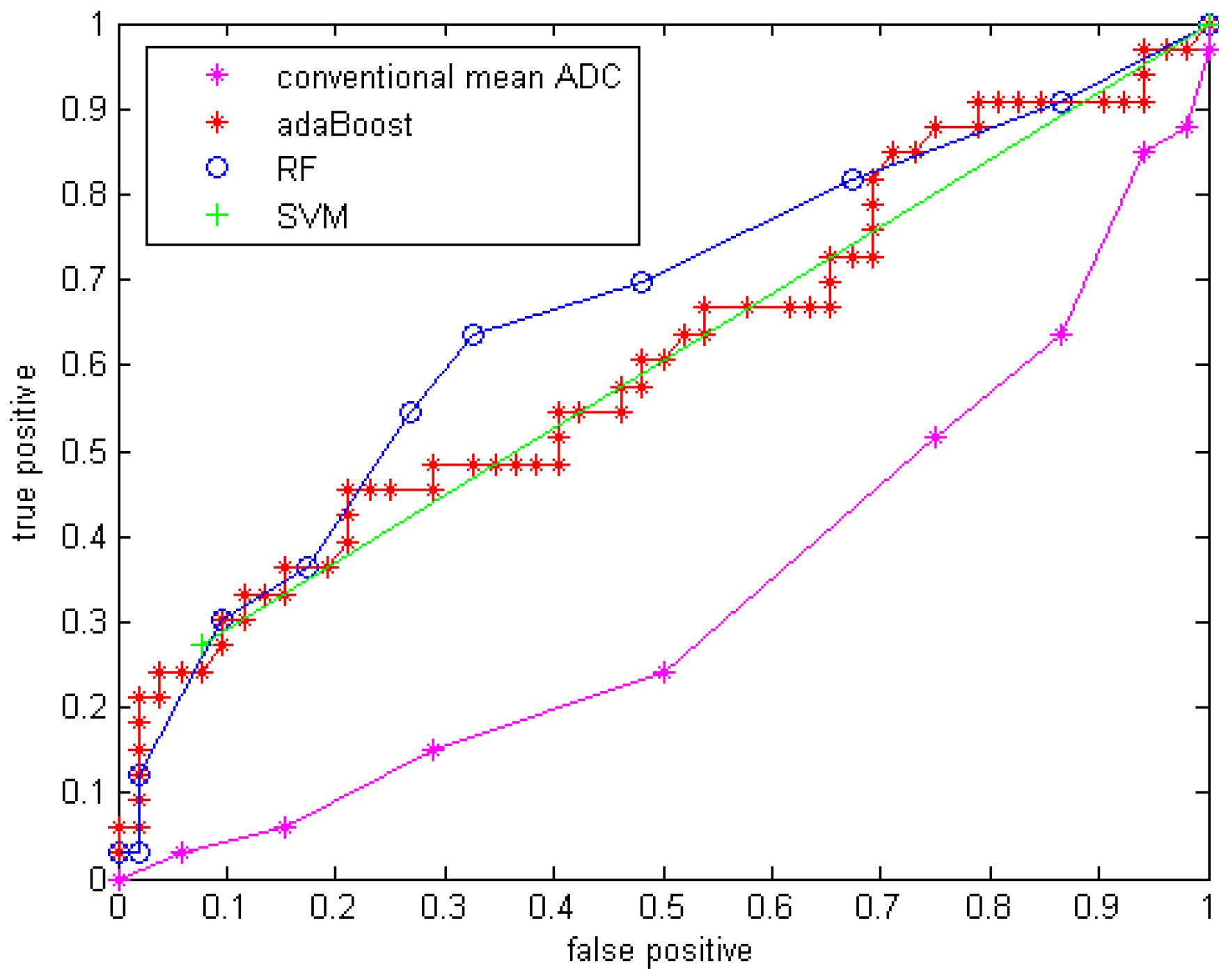
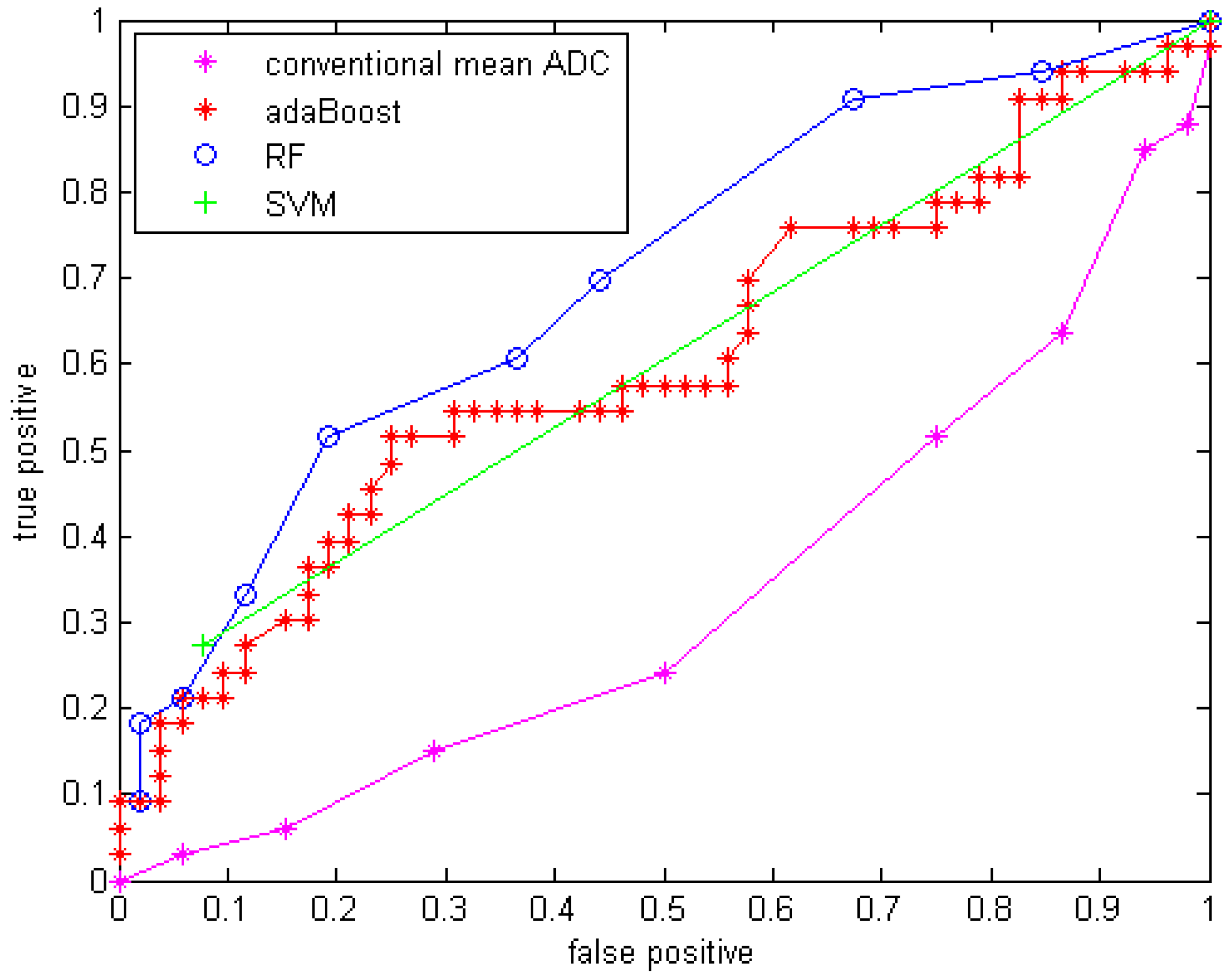
Compared to using only the mean ADC value, the quantitative statistical histogram features and the proposed classification system tremendously improved the accuracy from 29.4% to 69.41% (Az increased from 0.33 to 0.70). The statistical analysis indicates that all three classifiers are significantly different from the conventional mean ADC method with our dataset. Compared to general statistical histogram features, the classification with GMM features using random forest technique slightly improved the accuracy from 65.88% to 69.41%, while adaBoost and RF classifiers generated the same accuracy no matter whether GMM features were included. There is no significant difference between the three machine-learned classifiers.
The conventional mean ADC method performs worse than a random classifier (Az < 0.5). The reason is that conventionally researchers hypothesized that mean ADC increases because the tumor cell density decrease after an effective treatment. This assumption may not be valid for our dataset, because it involves in an anti-angiogenesis drug, which suppresses the cancer cell growth without necessary killing tumor cells (decreasing their density) at an early stage (5-7 weeks). Another possible reason is that in our dataset many of the GBM tumors are recurrent GBM tumors that are usually necrotic. The treatment tends to reduce necrosis and edema, which will diminish ADC. Essentially there are two competing processes at work: cell density, edema and necrosis [
25].
Another state-of-art study included features that capture spatial information in tumor heterogeneity features. Functional diffusion map (fDM) [
22,
23] is a popular technique studying the ADC value increase or decrease voxel-by-voxel. Moffat et al. applied fDM to 20 patients, classified patients into the three categories: PR, SD and PD, and reported 100% accuracy [
22]. However, the threshold they used for classification was determined from a single dataset of 20 patients used for both training and testing, while in our experiments, a cross validation analysis was performed. In Moffat et al’s study, they explored the assessment of fractionated radiation therapy for different types of brain tumors with 20 patients scanned on the same scanner [
22]. However, in our study, we focused on the GBM brain tumors treated by anti-angiogenesis drugs, which suppress the blood supply for the tumor cells and may not directly decrease the tumor cellularity. The difference in accuracy may come from the different mechanism of treatment. Additionally, our dataset is from GBM drug trials across multiple sites, thus our preliminary study is an important contribution for exploring DWI as an early imaging biomarker in a real pharmaceutical drug trial. In future work, we will extract texture feature to include spatial information, and shape features will be extracted as well. By introducing a new richer feature set indicating more useful tumor information, we aim to include more information about tumors and further improve the performance of the classification system.
One limitation of this study is that we classified CR, PR and SD as responders for the ground truth to achieve a binary classification. Since SD and PR may have different patterns in terms of their ADC histogram change, a multi-category classification system will be explored in future work. Another limitation of the study is that we used the Macdonald criteria at the eighth or tenth week after treatment for determining treatment response. In future work, time-to-progression and survival time will be a better endpoint to classify treatment response. Another limitation comes from the 3D ROI mapping tool. This tool is more computationally efficient compared to the co-registration techniques, but it cannot correct for patient motion. Therefore, in our study, a board-certified radiologist’s visually checked and edited all segmentation results as needed. In the future, a more sophisticated registration method with an image similarity measure may improve the accuracy of the tumor contours on ADC maps, and consequently improve the accuracy of the extracted features and the classifier performance.
ADC values obtained on pre-operative MRI scans are reported to be of prognostic value in patients with glioblastoma [
25,
42]. The term "prognosis" refers to predicting the likely outcome of treatment. ADC, reported to be inversely proportional to tumor cellularity, is gaining interest in predicting GBM tumor prognosis. Our proposed framework now uses changes in DW-MRI for early prediction of treatment response; however, the framework with feature extraction and machine learning technique could be generalized to pre-treatment DW-MRI for prognosis prediction.
In this study, we developed a CADrx framework with machine learning techniques to automatically predict tumor treatment response before the size change using DW-MRI. In our preliminary study, our major contributions are extracting statistical ADC histogram features, applying GMM to model the ADC histogram to interpret the competing effects of cellular density and edema, and applying machine learning techniques using all the extracted features. Cell density and edema may be reflected in ADC values before size changes are apparent on standard MRI sequences. Therefore, ADC holds promise as a biomarker, in determining both which tumors are more likely to respond to treatment and which tumors are actually responding.
In conclusion, this work shows that a CADrx system using quantitative ADC histogram features and a machine-learned classifier has better performance in treatment response assessment over conventional analysis using only a mean ADC value. This will have major implications for clinical trials. This work has potential clinical significance for early treatment response assessment in GBM.