Habitat Modeling of Alien Plant Species at Varying Levels of Occupancy
Abstract
:1. Introduction
2. Methods
2.1. Study Area

2.2. Species of Interest
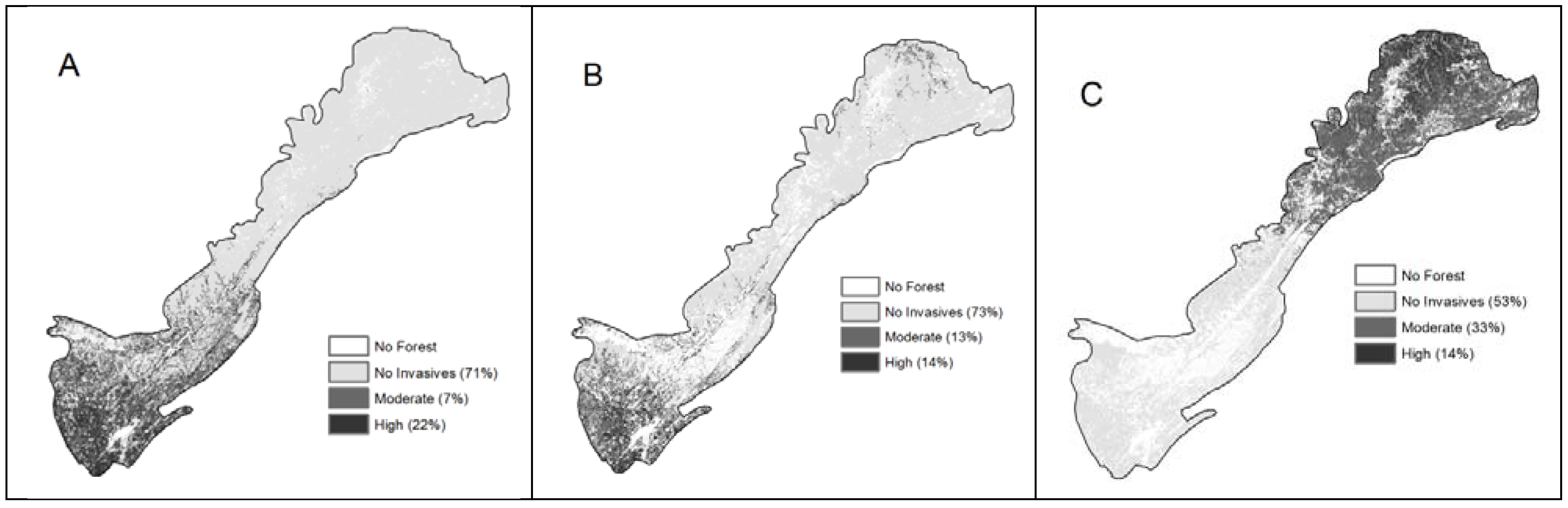
2.2.1. Privet
2.2.2. Tall Fescue
2.2.3. Silktree
2.3. Invasive Plant Occurrence
2.4. Landscape Variables
| Variable | Variable code | Citation | Res | Source | Mean | SD | Min | Max | |
|---|---|---|---|---|---|---|---|---|---|
| Landsat | Disturbance Index for 1975 | DI75 | [64] | 900 m2 | Landsat | 9.5 | 1.3 | −11.8 | 65.5 |
| Disturbance Index for 1990 | DI90 | [64] | 900 m2 | Landsat | −0.3 | 1.8 | −10.5 | 38.4 | |
| Disturbance Index for 2000 | DI00 | [64] | 900 m2 | Landsat | 0.0 | 2.0 | −9.8 | 48.1 | |
| Change in Disturbance Index between 1975 and 1990 | DI90-75 | [64] | 900 m2 | Landsat | |||||
| Change in Disturbance Index between 1990 and 2000 | DI00-90 | [64] | 900 m2 | Landsat | 0.4 | 2.2 | −40.7 | 59.9 | |
| NDVI in 1975 | NDVI75 | [65] | 900 m2 | Landsat | |||||
| NDVI 1990 | NDVI90 | [65] | 900 m2 | Landsat | 0.57 | 0.10 | −0.94 | 0.98 | |
| NDVI 2000 | NDVI00 | [65] | 900 m2 | Landsat | 0.45 | 0.15 | −0.96 | 0.99 | |
| Difference in NDVI between 1975 and 1990 | NDVI90-75 | [65] | 900 m2 | Landsat | |||||
| Difference in NDVI between 1990 and 2000 | NDVI00-90 | [65] | 900 m2 | Landsat | |||||
| Anthropogenic | Number of people per km2 in 2000 | CENSUS | [66] | Census block | Census 2000 TIGER | 24 | 46 | 3 | 2805 |
| Distance to road | RD_DIST | [66] | 900 m2 | Census 2000 TIGER | 397 | 375 | 0 | 3755 | |
| Density of roads within a km2 area in 2000 | RD_DEN | [66] | 900 m2 | Census 2000 TIGER | 1.3 | 1.0 | 0 | 15.6 | |
| Distance to major road | MRD_DIST | [66] | 900 m2 | Census 2000 TIGER | 5614 | 4717 | 0 | 26122 | |
| Residential in 2000 or 1990 within a 500 m buffer | RES ALL | [67] | 900 m2 | USGS LULC | |||||
| Residential presence within a 100 m buffer in 2000 | RES100 | [67] | 900 m2 | USGS LULC | 0.31 | 0.49 | 0 | 1 | |
| Residential presence within a 500 m buffer in 2000 | RES500 | [67] | 900m2 | USGS LULC | 0.71 | 0.49 | 0 | 1 | |
| Environmental | North | NORTH | [68] | 900 m2 | USGS NED | ||||
| East | EAST | [68] | 900 m2 | USGS NED | |||||
| Northness | NORTHNESS | [69] | 900 m2 | USGS NED | 0 | 0.18 | −0.88 | 0.83 | |
| Eastness | EASTNESS | [69] | 900 m2 | USGS NED | 0 | 0.19 | −0.83 | 0.84 | |
| Slope | SLOPE | [62] | 900 m2 | USGS NED | 12.9 | 8.9 | 0 | 62.3 | |
| Hillshade | HILL | [62] | 900 m2 | USGS NED | 237 | 17 | 59 | 254 | |
| Curvature | CURV | [62] | 900 m2 | USGS NED | |||||
| Elevation | DEM | [31] | 900 m2 | USGS NED | 383 | 168 | 0 | 1283 | |
| Climate | Average temperature from a 30-year average (1971–2000) | AVET | [32] | 900 m2 | PRISM | ||||
| Minimum temperature from a 30-year average (1971–2000) | MINT | [32] | 900 m2 | PRISM | 26.5 | 3.4 | 19 | 35 | |
| Maximum temperature from a 30-year average (1971–2000) | MAXT | [32] | 900 m2 | PRISM | |||||
| Average yearly rainfall from a 30-year average (1971–2000) | RAIN | [32] | 900 m2 | PRISM | 54 | 5 | 41 | 75 | |
| Land Cover | Change in forest between 2000 and 1990 within a 100-m buffer | FC100 | [67] | 900 m2 | USGS LULC | ||||
| Change in forest between 2000 and 1990 within a 500-m buffer | FC500 | [67] | 900 m2 | USGS LULC | 0.12 | 0.13 | −1 | 0.99 | |
| Proportion of forest in 2000 with in a 100-m buffer | F00 100 | [67] | 900 m2 | USGS LULC | 0.90 | 0.17 | 0.03 | 1 | |
| Proportion of forest in 2000 with in a 500-m buffer | F00 500 | [67] | 900 m2 | USGS LULC | |||||
| Proportion of farming in 2000 with in a 100-m buffer | FARM100 | [67] | 900 m2 | USGS LULC | |||||
| Proportion of farming in 2000 with in a 500-m buffer | FARM500 | [67] | 900 m2 | USGS LULC | 0.07 | 0.13 | 0 | 0.98 | |
| Categorical land use in 1990 based on Andersons groupings | LULC90 | [67] | 900 m2 | USGS LULC | Categorical | ||||
| Categorical land use in 2000 based on Andersons groupings | LULC00 | [67] | 900 m2 | USGS LULC | Categorical | ||||
| Water | Distance from a stream | RIV DIS | [70] | 900 m2 | USGS | 336 | 267 | 0 | 3288 |
| Density of streams within a km2 area | RIV_DEN | [70] | 900 m2 | USGS | 0.96 | 0.51 | 0 | 6.65 | |
| Occurrence of a wetland or stream within 100 m | WATER100 | [67] | 900 m2 | USGS LULC | 0.05 | 0.51 | 0 | 1 | |
| Occurrence of a wetland or stream within 500 m | WATER500 | [67] | 900 m2 | USGS LULC | 0.30 | 0.50 | 0 | 1 | |
2.5. Models
2.6. Data Selection
| Training | Test | |||
|---|---|---|---|---|
| Occurrence | Absence | Occurrence | Absence | |
| Privet | 200 (10.4%) | 1125 (59.0%) | 100 (5.2%) | 482 (25.4%) |
| Tall fescue | 65 (3.4%) | 1270 (66.6%) | 28 (1.5%) | 544 (28.5%) |
| Silktree | 31 (1.6%) | 1304 (68.4%) | 13 (0.7%) | 559 (29.3%) |
3. Results and Discussion
| Species | Model | Group | Threshold | Omission rate | AUC | ||
|---|---|---|---|---|---|---|---|
| Train | Test | Train | Test | ||||
| Privet | L | Landsat | 0.18 | 0.07 | 0.10 | 0.70 | 0.66 |
| M | Landsat | 0.47 | 0.25 | 0.37 | 0.74 | 0.68 | |
| L | Anthro | 0.16 | 0.06 | 0.08 | 0.76 | 0.72 | |
| M | Anthro | 0.33 | 0.09 | 0.20 | 0.77 | 0.70 | |
| L | Enviro | 0.16 | 0.04 | 0.05 | 0.84 | 0.82 | |
| M | Enviro | 0.34 | 0.10 | 0.10 | 0.80 | 0.80 | |
| L | Climate | 0.16 | 0.20 | 0.25 | 0.83 | 0.82 | |
| M | Climate | 0.42 | 0.18 | 0.30 | 0.83 | 0.82 | |
| L | Land use | 0.13 | 0.05 | 0.11 | 0.83 | 0.82 | |
| M | Land use | 0.30 | 0.10 | 0.13 | 0.81 | 0.79 | |
| L | Water | 0.17 | 0.10 | 0.21 | 0.66 | 0.65 | |
| M | Water | 0.52 | 0.51 | 0.51 | 0.66 | 0.67 | |
| L | Composite | 0.17 | 0.02 | 0.05 | 0.91 | 0.89 | |
| M | Composite | 0.28 | 0.07 | 0.14 | 0.86 | 0.83 | |
| Tall Fescue | L | Landsat | 0.06 | 0.32 | 0.54 | 0.74 | 0.65 |
| M | Landsat | 0.45 | 0.34 | 0.64 | 0.76 | 0.61 | |
| L | Anthro | 0.05 | 0.41 | 0.40 | 0.65 | 0.63 | |
| M | Anthro | 0.40 | 0.26 | 0.32 | 0.70 | 0.67 | |
| L | Enviro | 0.05 | 0.49 | 0.38 | 0.60 | 0.62 | |
| M | Enviro | 0.49 | 0.34 | 0.50 | 0.75 | 0.66 | |
| L | Climate | 0.06 | 0.28 | 0.30 | 0.73 | 0.70 | |
| M | Climate | 0.44 | 0.15 | 0.10 | 0.77 | 0.84 | |
| L | Land use | 0.06 | 0.36 | 0.42 | 0.66 | 0.59 | |
| M | Land use | 0.46 | 0.24 | 0.39 | 0.72 | 0.60 | |
| L | Water | No Model | |||||
| M | Water | 0.47 | 0.20 | 0.32 | 0.61 | 0.54 | |
| L | Composite | 0.05 | 0.25 | 0.21 | 0.78 | 0.75 | |
| M | Composite | 0.42 | 0.25 | 0.35 | 0.82 | 0.75 | |
| Tall Fescue | L | Landsat | 0.06 | 0.32 | 0.54 | 0.74 | 0.65 |
| M | Landsat | 0.47 | 0.29 | 0.36 | 0.73 | 0.73 | |
| L | Anthro | 0.02 | 0.22 | 0.41 | 0.75 | 0.73 | |
| M | Anthro | 0.47 | 0.29 | 0.21 | 0.84 | 0.90 | |
| L | Enviro | 0.02 | 0.10 | 0.15 | 0.83 | 0.80 | |
| M | Enviro | 0.30 | 0.06 | 0.29 | 0.82 | 0.78 | |
| L | Climate | 0.02 | 0.19 | 0.17 | 0.77 | 0.75 | |
| M | Climate | 0.42 | 0.13 | 0.07 | 0.77 | 0.82 | |
| L | Land use | 0.02 | 0.35 | 0.37 | 0.81 | 0.85 | |
| M | Land use | 0.42 | 0.25 | 0.43 | 0.80 | 0.70 | |
| L | Water | 0.02 | 0.29 | 0.35 | 0.74 | 0.75 | |
| M | Water | 0.49 | 0.32 | 0.28 | 0.76 | 0.76 | |
| L | Composite | 0.02 | 0.16 | 0.38 | 0.89 | 0.80 | |
| M | Composite | 0.27 | 0.06 | 0.07 | 0.91 | 0.90 | |
| Species | Privet | Tall fescue | Silktree | ||||
|---|---|---|---|---|---|---|---|
| Model | L | M | L | M | L | M | |
| Landsat | DI00 | (+)6 | |||||
| Anthropogenic | CENSUS | (+)24 | |||||
| RD DEN | (+)4 | (+)15 | (+)35 | (+)16 | |||
| RD DIST | (−)3 | ||||||
| MRD DIST | (−)3 | ||||||
| RES100 | (−)8 | ||||||
| Environmental | DEM | (−)13 | (−)7 | (∩)19 | (−)58 | (−)48 | |
| NORTHNESS | (−)30 | (−)7 | |||||
| SLOPE | (−)6 | ||||||
| Climatic | MINT | (+)66 | (+)55 | (−)62 | (−)54 | ||
| RANN | (U)5 | (∩)10 | |||||
| Land use | F00 100 | (−)10 | |||||
| FARM500 | (+)4 | (∩)10 | |||||
| LULC90 | 7 | ||||||
| Water | WATER500 | (−)1 | (+)12 | ||||
| Proportion forest area invaded | 24% | 28% | 46% | 16% | 20% | 21% | |
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Hulme, P.E.; Pyšek, P.; Nentwig, W.; Vila, M. Will threat of biological invasions unite the European Union? Science 2009, 324, 40–41. [Google Scholar]
- Vilà, M.; Basnou, C.; Pyšek, P.; Josefsson, M.; Genovesi, P.; Gollasch, S.; Nentwig, W.; Olenin, S.; Roques, A.; Roy, D.; et al. How well do we understand the impacts of alien species on ecosystem services? A pan-European, cross-taxa assessment. Front. Ecol. Environ. 2010, 8, 135–144. [Google Scholar] [CrossRef] [Green Version]
- Mainka, S.A.; Howard, G.W. Climate change and invasive species: Double jeopardy. Integr. Zool. 2010, 5, 102–111. [Google Scholar] [CrossRef]
- Ricciardi, A. Are modern biological invasions an unprecedented form of global change? Conserv. Biol. 2007, 21, 329–336. [Google Scholar] [CrossRef]
- Vitousek, P.M.; D’Antonio, C.M.; Loope, L.L.; Rejmanek, M.; Westbrooks, R. Introduced species: A significant component of human-caused global change. N. Z. J. Ecol. 1997, 21, 1–16. [Google Scholar]
- Guisan, A.; Thuiller, W. Predicting species distribution: Offering more than simple habitat models. Ecol. Lett. 2005, 8, 993–1009. [Google Scholar] [CrossRef]
- Smolik, M.G.; Dullinger, S.; Essl, F.; Kleinbauer, I.; Leitner, M.; Peterseil, J.; Stadler, L.M.; Vogl, G. Integrating species distribution models and interacting particle systems to predict the spread of an invasive alien plant. J. Biogeogr. 2010, 37, 411–422. [Google Scholar] [CrossRef]
- Guisan, A.; Lehmann, A.; Ferrier, S.; Austin, M.; Overton, J.M.C.; Aspinall, R.; Hastie, T. Making better biogeographical predictions of species’ distributions. J. Appl. Ecol. 2006, 43, 386–392. [Google Scholar] [CrossRef]
- Kearney, M.R.; Wintle, B.A.; Porter, W.P. Correlative and mechanistic models of species distribution provide congruent forecasts under climate change. Conserv. Lett. 2010, 3, 203–213. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Thuiller, W.; Miaud, C. Prediction and validation of the potential global distribution of a problematic non-native invasive species; the American bullfrog. Divers. Distrib. 2002, 8, 49–56. [Google Scholar] [CrossRef]
- Hoffman, J.D.; Aguilar-Amuchastegui, N.; Tyre, A.J. Use of simulated data from a process-based habitat model to evaluate methods for predicting species occurrence. Ecography 2010, 33, 656–666. [Google Scholar]
- Kumar, S.; Stohlgren, T.J.; Chong, G.W. Spatial heterogeneity influences native and alien plant species richness. Ecology 2006, 87, 3186–3199. [Google Scholar]
- Stohlgren, T.J.; Binkley, D.; Chong, G.W.; Kalkhan, M.A.; Schell, L.D.; Bull, K.A.; Otsuki, Y.; Newman, G.; Bashkin, M.; Son, Y. Exotic plant species invade hot spots of native plant diversity. Ecol. Monogr. 1999, 69, 25–46. [Google Scholar] [CrossRef]
- With, K.A.; Crist, T.O. Critical thresholds in species’ responses to landscape structure. Ecology 1995, 76, 2446–2459. [Google Scholar] [CrossRef]
- Pickett, S.T.A.; Cadenasso, M.L. Landscape ecology: Spatial heterogeneity in ecological systems. Science 1995, 269, 331–334. [Google Scholar]
- Wagner, H.H.; Fortin, M.J. Spatial analysis of landscapes: Concepts and statistics. Ecology 2005, 86, 1975–1987. [Google Scholar] [CrossRef]
- Collingham, Y.C.; Wadsworth, R.A.; Huntley, B.; Hulme, P.E. Predicting the spatial distribution of non-indigenous riparian weeds: Issues of spatial scale and extent. J. Appl. Ecol. 2000, 37, 13–27. [Google Scholar] [CrossRef]
- Lemke, D.; Hulme, P.E.; Brown, J.A.; Tadesse, W. Distribution modelling of Japanese honeysuckle (Lonicera japonica) invasion in the Cumberland Plateau and Mountain Region, USA. For. Ecol. Manag. 2011, 262, 139–149. [Google Scholar] [CrossRef]
- Robertson, M.P.; Villet, M.H.; Palmer, A.R. A fuzzy classification technique for predicting species’ distributions: Application using invasive alien plants and indigenous insects. Divers. Distrib. 2004, 10, 461–474. [Google Scholar] [CrossRef]
- Underwood, E.C.; Klinger, R.; Moore, P.E. Predicting patterns of non-native plant invasions in Yosemite National Park, California, USA. Divers. Distrib. 2004, 10, 447–459. [Google Scholar] [CrossRef]
- Hoffman, J.D.; Narumalani, S.; Mishra, D.R.; Merani, P.; Wilson, R.G. Predicting potential occurrence and spread of invasive plant species along the North Platte River, Nebraska. Invasive Plant Sci. Manag. 2008, 1, 359–367. [Google Scholar] [CrossRef]
- Dullinger, S.; Kleinbauer, I.; Peterseil, J.; Smolik, M.; Essl, F. Niche based distribution modelling of an invasive alien plant: Effects of population status, propagule pressure and invasion history. Biol. Invasions 2009, 11, 2401–2414. [Google Scholar] [CrossRef]
- Araujo, M.B.; New, M. Ensemble forecasting of species distributions. Trends in Ecol. Evol. 2007, 22, 42–47. [Google Scholar] [CrossRef]
- Stohlgren, T.J.; Ma, P.; Kumar, S.; Rocca, M.; Morisette, J.T.; Jarnevich, C.S.; Benson, N. Ensemble habitat mapping of invasive plant species. Risk Anal. 2010, 30, 224–235. [Google Scholar] [CrossRef]
- Smalley, G.W. Classification and Evaluation of Forest Sites on the Southern Cumberland Plateau; General Technical Report Southern-23; U.S. Department of Agriculture, Forest Service, Southern Forest Experiment Station: New Orleans, LA, USA, 1979. [Google Scholar]
- Smalley, G.W. U.S. Department of Agriculture, Forest Service, Southern Forest Experiment Station; U.S. Department of Agriculture, Forest Service, Southern Forest Experiment Station: New Orleans, LA, USA, 1982. [Google Scholar]
- Smalley, G.W. Classification and Evaluation of Forest Sites in the Cumberland Mountains; General Technical Report Southern-50; U.S. Department of Agriculture, Forest Service, Southern Forest Experiment Station: New Orleans, LA, USA, 1984. [Google Scholar]
- Smalley, G.W. Classification and Evaluation of Forest Sites on the Northern Cumberland Plateau; General Technical Report Southern-60; U.S. Department of Agriculture, Forest Service, Southern Forest Experiment Station: New Orleans, LA, USA, 1986. [Google Scholar]
- Ricketts, T.H.; Dinerstein, E.; Olson, D.M.; Loucks, C.J.; Eichbaum, W. Terrestrial Ecoregions of North America: A Conservation Assessment; Island Press: Washington, DC, USA, 1999. [Google Scholar]
- Homer, C.; Huang, C.; Yang, L.; Wylie, B. Development of a 2001 national land cover database for the United States. Photogramm. Eng. Remote Sens. 2004, 70, 829–840. [Google Scholar]
- Gesch, D.; Oimoen, M.; Greenlee, S.; Nelson, C.; Steuck, M.; Tyler, D. The national elevation dataset. Photogramm. Eng. Remote Sens. 2002, 68, 5–11. [Google Scholar]
- PRISM Group. 30-year average (1971–2000) PRISM data. Oregon State University: Corvallis, OR, USA. Available online: http://www.prismclimate.org (accessed on 9 December 2007).
- Wear, D.N.; Greis, J.G. Southern forest resource assessment: Summary of findings. J. For. 2002, 100, 6–14. [Google Scholar]
- McGrath, D.A; Evans, J.P.; Smith, C.K.; Haskell, D.G.; Pelkey, N.W.; Gottfried, R.R.; Brockett, C.D.; Lane, M.D.; Williams, E.D. Mapping land-use change and monitoring the impacts of hardwood-to-pine conversion on the Southern Cumberland Plateau in Tennessee. Earth Interact. 2004, 8, 1–24. [Google Scholar]
- U.S. Department of Agriculture, Forest Service, The Forest Inventory and Analysis Database: Database Description and Users’ Guide, Version 3.0; USDA FS: Washington, DC, USA, 2007.
- U.S. Department of Agriculture (USDA) Plants. National Plants Database. USDA: National Plant Data Team, Greensboro, NC, USA, 2011. Available online: http://plants.usda.gov (accessed on 10 April 2011).
- Dirr, M.A. Manual of Woody Landscape Plants: Their Identification, Ornamental Characteristics, Culture, Propagation and Uses; Stipes Publishing: Champaign, IL, USA, 1998. [Google Scholar]
- Maddox, V.; Byrd, J.; Serviss, B. Identification and control of invasive privets (Ligustrum spp.) in the middle southern United States. Invasive Plant Sci. Manag. 2010, 3, 482–488. [Google Scholar] [CrossRef]
- Miller, J.H.; Chambliss, E.B.; Loewenstein, N.J. A Field Guide for the Identification of Invasive Plants in Southern Forests; General Technical Report Southern Research Station-119; U.S. Department of Agriculture, Forest Service, Southern Research Station: Asheville, NC, USA, 2010. [Google Scholar]
- Merriam, R.W.; Feil, E. The potential impact of an introduced shrub on native plants diversity and forest regeneration. Biol. Invasions 2002, 4, 369–373. [Google Scholar] [CrossRef]
- Wilcox, J.; Beck, C.W. Effects of Ligustrum sinense Lour. (Chinese privet) on abundance and diversity of songbirds and native plants in a southeastern nature preserve. Southeast. Nat. 2007, 6, 535–550. [Google Scholar] [CrossRef]
- Hanula, J.L.; Horn, S.; Taylor, J.W. Chinese privet (Ligustrum sinense) removal and its effect on native plant communities of riparian forests. Invasive Plant Sci. Manag. 2009, 2, 292–300. [Google Scholar] [CrossRef]
- Shelton, M.G.; Cain, M.D. Potential carry-over of seeds from 11 common shrub and vine competitors of loblolly and shortleaf pines. Can. J. For. Res. 2002, 32, 412–419. [Google Scholar] [CrossRef]
- Greenberg, C.H.; Walter, S.T. Fleshy fruit removal and nutritional composition of winter-fruiting plants: A comparison of non-native invasive and native species. Nat. Areas J. 2010, 30, 312–321. [Google Scholar] [CrossRef]
- Stromayer, K.; Warren, R.J.; Johnson, A.S.; Hale, P.E.; Rogers, C.L.; Tucker, C.L. Chinese privet and the feeding ecology of white-tailed deer: The role of an exotic plant. J. Wildl. Manag. 1998, 62, 1321–1329. [Google Scholar] [CrossRef]
- Hannaway, D.; Fransen, S.; Cropper, J.; Teel, M.; Chaney, M.; Griggs, T.; Halse, R.; Hart, J.; Cheeke, P. Tall Fescue (Festuca arundinacea Schreb). A Pacific Northwest Extension Publication PWN 504. Oregon State University: Corvallis, OR, USA, 1999. [Google Scholar]
- Fleming, C.A.; Wofford, B.E. The vascular flora of Fall Creek Falls State Park, Van Buren and Bledsoe Counties, Tennesse. Castanea 2004, 69, 164–184. [Google Scholar] [CrossRef]
- Pedersen, J.F.; Lacefield, G.D.; Ball, D.M. A review of the agronomic characteristics of endophyte-free and endophyte-infected tall fescue. Appl. Agric. Res. 1990, 3, 188–194. [Google Scholar]
- Spyreas, G.; Gibson, D.J.; Middleton, B.A. Effects of endophyte infection in tall fescue (Festuca arundinacea, Poaceae) on community diversity. Int. J. Plant Sci. 2001, 162, 1237–1245. [Google Scholar] [CrossRef]
- Clay, K.; Schardl, C. Evolutionary Origins and Ecological Consequences of Endophyte Symbiosis with Grasses; University Chicago Press: Chicago, IL, USA, 2002. [Google Scholar]
- Schardl, C.L.; Leuchtmann, A.; Spiering, M.J. Symbioses of grasses with seedborne fungal endophytes. Annu. Rev. Plant Biol. 2004, 55, 315–340. [Google Scholar] [CrossRef]
- Rudgers, J.; Clay, K. An invasive plant-fungal mutualism reduces arthropod diversity. Ecol. Lett. 2008, 11, 831–840. [Google Scholar] [CrossRef]
- Creager, R.A. Seed Germination, physical and chemical control of catclaw mimosa (Mimosa pigra var. pigra). Weed Technol. 1992, 6, 884–891. [Google Scholar]
- Ares, A.; Burner, D.M.; Brauer, D.K. Soil phosphorus and water effects on growth, nutrient and carbohydrate concentrations, d13C, and nodulation of silktree (Albizia julibrissin Durz.) on a highly weathered soil. Agrofor. Syst. 2009, 76, 317–325. [Google Scholar]
- Addlestone, B.J.; Mueller, J.P.; Luginbuhl, P.M. The establishment and early growth of three leguminous tree species for use in silvopastoral systems in the southern USA. Agrofor. Syst. 1998, 44, 253–265. [Google Scholar] [CrossRef]
- Bransby, D.I.; Sladden, S.E.; Aiken, G.E. American Forage Grass Council. Silktree as a Forage Plant: A Preliminary Evaluation. In Proceedings of the Forage Grassland Conference, Georgetown, TX, USA, 5 April 1992; 1, pp. 28–31.
- Matta-Machado, R.P.; Jordan, C.F. Nutrient dynamics during the first three years of an alley cropping agroecosystem in southern USA. Agrofor. Syst. 1995, 30, 351–362. [Google Scholar] [CrossRef]
- Rhoades, C.C.; Nissen, T.M.; Kettler, J.S. Soil nitrogen dynamics in alley cropping and no-till systems on ultisols of the Georgia Piedmont, USA. Agrofor. Syst. 1997, 39, 31–44. [Google Scholar] [CrossRef]
- Jordan, C.F. Organic farming and agroforestry: Alley cropping for mulch production for organic farms in southern United States. Agrofor. Syst. 2004, 61, 79–90. [Google Scholar] [CrossRef]
- Loewenstein, N.J.; Loewenstein, E.F. Alien plants in the understory of riparian forests across a land use gradient in the Southeast. Urban Ecosyst. 2005, 8, 79–91. [Google Scholar] [CrossRef]
- Birdsey, R.A.; Schreuder, H.T. An Overview of Forest Inventory and Analysis Estimation Procedures in the Eastern United States–with an Emphasis on the Components of Change; USDA Forest Service, Rocky Mountain Forest and Range Experiment Station: Fort Collins, CO, USA, 1992. [Google Scholar]
- Environmental Systems Research Institute (ESRI), ArcGIS; Environmental Systems Research Institute: Redlands, CA, USA, 2009.
- Earth Resources Data Analysis System (ERDAS IMAGINE 9.2.); Intergraph Corporation: Norcross, GA, USA, 2008.
- Healey, S.P.; Cohen, W.B.; Yang, Z.Q.; Krankina, O.N. Comparison of tasseled cap-based Landsat data structures for use in forest disturbance detection. Remote Sens. Environ. 2005, 97, 301–310. [Google Scholar]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- United States Bureau of the Census (USBOC), Tiger Files; USBOC, Geography Division: Washington, DC, USA, 2000.
- Anderson, J.R.; Hardy, E.E.; Roach, J.T.; Witmer, R.E. A Land Use and Land Cover Classification System for Use with Remote Sensor Data. In United States Geological Survey Professional Paper 964; United States Government Printing Office: Washington, DC, USA, 1976. [Google Scholar]
- Guisan, A.; Weiss, S.B.; Weiss, A.D. GLM vs. CCA spatial modeling of plant species distribution. Plant Ecol. 1999, 143, 107–122. [Google Scholar] [CrossRef]
- Piedallu, C.; Gegout, J. Efficient assessment of topographic solar radiation to improve plant distribution models. Agric. For. Meteorol. 2008, 148, 1696–1706. [Google Scholar] [CrossRef]
- Simley, J.D.; Carswell, W.J., Jr. The National Map—Hydrography; U.S. Geological Survey Fact Sheet 2009-3054; U.S. Geological Survey: Reston, VA, USA, 2009. [Google Scholar]
- Hosmer, D.W.; Lemeshow, S. Applied Logistic Regression; Wiley Interscience: New York, NY, USA, 2000. [Google Scholar]
- Phillips, S.; Anderson, R.; Schapire, R. Maximum entropy modelling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- SAS, Version 9.2; SAS Institute: Cary, FL, USA, 2009.
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Manel, S.; Williams, H.C.; Ormerod, S.J. Evaluating presence-absence models in ecology: The need to account for prevalence. J. Appl. Ecol. 2002, 38, 921–931. [Google Scholar] [CrossRef]
- Lobo, J.M.; Jiménez-Valverde, A. AUC: A misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 2008, 17, 145–151. [Google Scholar] [CrossRef]
- Oommen, T.; Baise, L.G.; Vogel, R.M. Sampling bias and class imbalance in maximum-likelihood logistic regression. Math. Geosci. 2010, 43, 99–120. [Google Scholar]
- Wisz, M.S.; Hijmanss, R.J.; Peterson, A.T.; Graham, C.H.; Guisan, A. Effects of sample size on the performance of species distribution models. Divers. Distrib. 2008, 14, 763–773. [Google Scholar] [CrossRef]
- Peterson, A.T. Predicting the geography of species’ invasions via ecological niche modelling. Q. Rev. Biol. 2003, 78, 419–433. [Google Scholar] [CrossRef]
- Turner, W.; Spector, S.; Gardiner, N.; Fladeland, M.; Sterling, E.; Steininger, M. Remote sensing for biodiversity science and conservation. Trends Ecol. Evol. 2003, 18, 306–314. [Google Scholar] [CrossRef]
- Sester, M. Optimization approaches for generalization and data abstraction. Int. J. Geogr. Inf. Sci. 2005, 19, 871–897. [Google Scholar] [CrossRef]
- Barbosa, A.M.; Real, R.; Vargas, J.M. Transferability of environmental favourability models in geographic space: The case of the Iberian desman (Galemys pyrenaicus) in Portugal and Spain. Ecol. Model. 2009, 220, 747–754. [Google Scholar] [CrossRef]
- Ducheyne, E.I.; De Wulf, R.R.; De Baets, B. A spatial approach to forest-management optimization: Linking GIS and multiple objective genetic algorithms. Int. J. Geogr. Inf. Sci. 2006, 20, 917–928. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lemke, D.; Brown, J.A. Habitat Modeling of Alien Plant Species at Varying Levels of Occupancy. Forests 2012, 3, 799-817. https://doi.org/10.3390/f3030799
Lemke D, Brown JA. Habitat Modeling of Alien Plant Species at Varying Levels of Occupancy. Forests. 2012; 3(3):799-817. https://doi.org/10.3390/f3030799
Chicago/Turabian StyleLemke, Dawn, and Jennifer A. Brown. 2012. "Habitat Modeling of Alien Plant Species at Varying Levels of Occupancy" Forests 3, no. 3: 799-817. https://doi.org/10.3390/f3030799




