Assessment of Low Density Full-Waveform Airborne Laser Scanning for Individual Tree Detection and Tree Species Classification
Abstract
:1. Introduction
2. Study Area and Material
2.1. Study Area

2.2. Field Measurements
| Tree species | Min. | Max. | Mean | Standard deviation | Number of trees | |
|---|---|---|---|---|---|---|
| Tree height (m) | Pine | 4.3 | 35.2 | 16.8 | 5.7 | 2613 |
| Spruce | 4.2 | 34.2 | 18.6 | 7.1 | 1726 | |
| Deciduous | 3.6 | 33.0 | 15.4 | 5.7 | 1193 | |
| DBH (cm) | Pine | 7.0 | 63.1 | 18.1 | 7.8 | |
| Spruce | 7.0 | 69.8 | 16.0 | 8.7 | ||
| Deciduous | 7.0 | 59.5 | 13.5 | 6.8 | ||
| Plot density (trees/ha) | 31.8 | 2037.2 | 603.0 | 388.6 |
2.3. ALS Data
3. Methods
3.1. Full Waveform Decomposition and Features
- N: Number of peaks extracted from the waveform;
- E: Sum of the waveform samples above the noise level;
- A_i: peak amplitude for the first four returns (peaks), i = 1, 2, 3, 4;
- W_i: number of samples that constitute the peak for the first four returns (peaks), i = 1, 2, 3, 4;
- R: waveform range calculated as the Euclidean distance between the first and the last peaks that was extracted from the waveform. This is shown as a blue line at the bottom of the top row waveform pictures in Figure 2;
- DB_i: distance between first above noise point and the point at which 50%, 80% and 95% of the energy is received (cumulative sum of amplitudes), i = 50, 80, 95;
- DA_i: Above ground height for 50%, 80% and 95% of the total waveform (cumulative sum of amplitudes), i = 50, 80, 95.

3.2. Individual Tree Detection
3.3. ALS-Derived Features for Trees
| Index | Feature | Description |
|---|---|---|
| DSC Features | ||
| 1 | P | Pulse penetration as the ratio of ground hit to total hits |
| 2 | mH | Arithmetic mean of heights |
| 3 | sH | Standard deviation of heights |
| 4 | rH | Range of height |
| 5 | CA | Crown area as the area of convex hull in 2D |
| 6 | CV | Crown volume as the convex hull in 3D |
| 7–16 | H_i | 0th to 90th percentile of canopy height distribution with a interval of 10% |
| 17 | maxH | Height Maximum |
| 18–26 | DS_i | Percentage of returns below 10%–90% of total height with a interval of 10% |
| 27 | MaxD | Maximum crown diameter when crown was considered an ellipse |
| FWF Features | ||
| 28–44 | Mean (X) | Average of all FWF hitting a tree for each feature described in Section 3.1, where X = {N, E, A_i, W_i, R, DB_i, DA_i} |
| 45–61 | Max (X) | Maximum of all FWF hitting a tree for each feature described in Section 3.1, where X = {N, E, A_i, W_i, R, DB_i, DA_i} |
| 62–78 | Std (X) | Standard deviation of all FWF hitting a tree for each feature described in Section 3.1, where X = {N, E, A_i, W_i, R, DB_i, DA_i} |
3.4. Random Forest for Feature Selection and Classification
3.5. Accuracy Assessment



4. Results
4.1. FWF(Full-Waveform) Decomposition

4.2. Individual Tree Detection



| No. of detected trees | Nt | Nc | No | r (%) | p (%) | F (%) | |
|---|---|---|---|---|---|---|---|
| DSC point data | 3362 | 2895 | 467 | 2637 | 52.3 | 86.1 | 65.1 |
| FWF point data | 3695 | 3030 | 665 | 2502 | 54.8 | 82.0 | 65.7 |
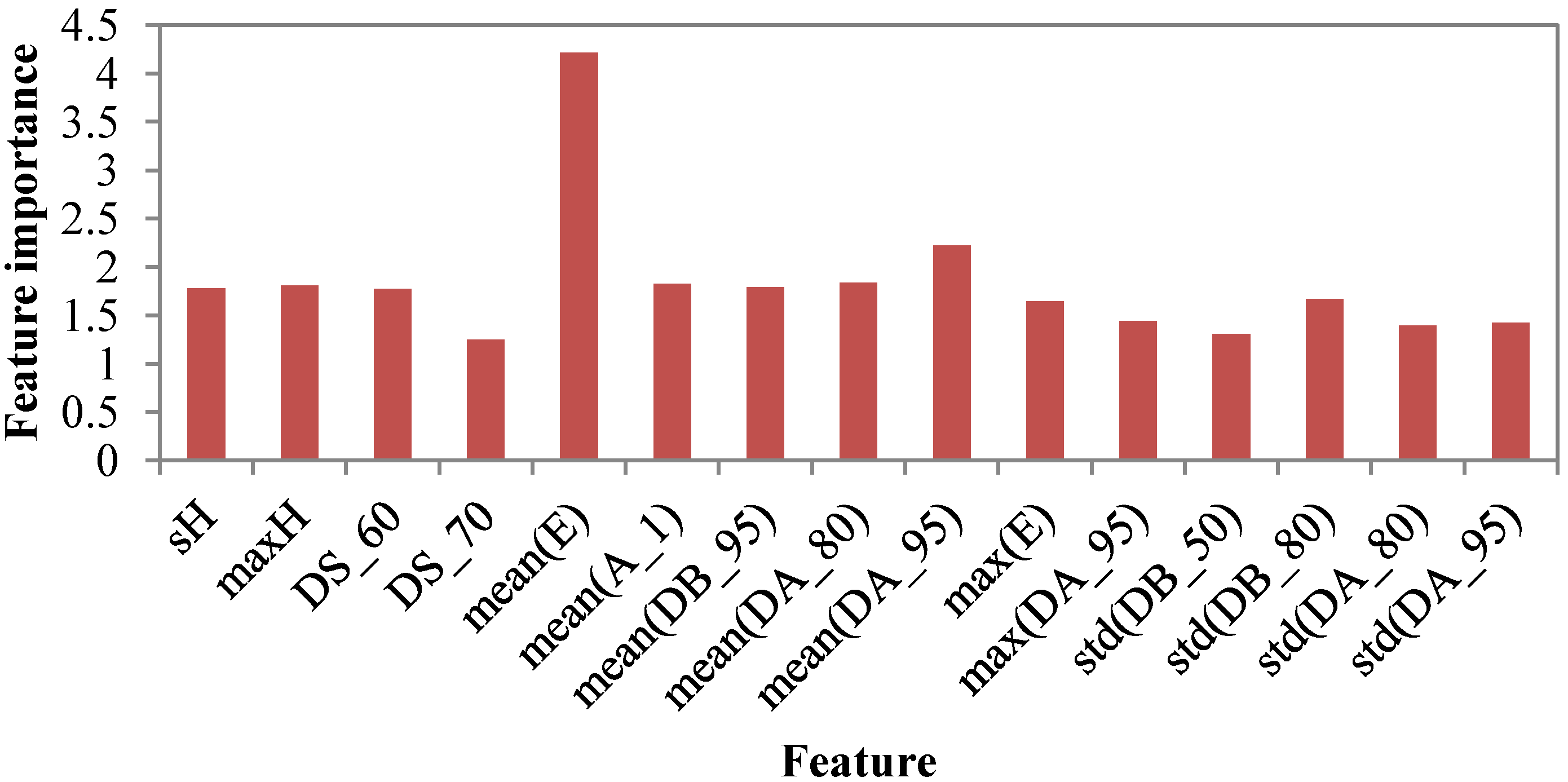
4.3. Feature Importance

4.4. Tree Species Classification
| Predicted class | |||||||
|---|---|---|---|---|---|---|---|
| With DSC features | With DSC and FWF features | ||||||
| Pine | Spruce | Birch | Pine | Spruce | Birch | ||
| Reference class | Pine | 1284 | 139 | 156 | 1362 | 122 | 95 |
| Spruce | 235 | 312 | 116 | 186 | 367 | 110 | |
| Birch | 339 | 113 | 201 | 168 | 90 | 395 | |
| With DSC Features | With DSC and FWF features | |||||
|---|---|---|---|---|---|---|
| Producer’s accuracy | User’s accuracy | Overall accuracy | Producer’s accuracy | User’s accuracy | Overall accuracy | |
| Pine | 81.3 | 69.1 | 86.3 | 79.4 | ||
| Spruce | 47.1 | 55.3 | 55.4 | 63.4 | ||
| Birch | 30.8 | 42.5 | 60.5 | 65.8 | ||
| 62.1 | 73.4 | |||||
| Predicted class | |||||
|---|---|---|---|---|---|
| Pine | Spruce | Birch | Producer’s accuracy (%) | ||
| Reference class | Pine | 1356 | 123 | 100 | 85.9 |
| Spruce | 217 | 337 | 109 | 50.8 | |
| Birch | 185 | 91 | 377 | 57.7 | |
| User’s accuracy (%) | 77.1 | 61.2 | 64.3 | overall = 71.5 | |
5. Discussion
5.1. Individual Tree Detection
5.2. Tree Species Classification
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Li, W.; Guo, Q.; Jakubowski, M.K.; Kelly, M. A new method for segmenting individual trees from the LiDAR point cloud. Photogramm. Eng. Remote Sens. 2012, 78, 75–84. [Google Scholar] [CrossRef]
- Kankare, V.; Räty, M.; Yu, X.; Holopainen, M.; Vastaranta, M.; Kantola, T.; Hyyppä, J.; Hyyppä, H.; Alho, P.; Viitala, R. Single tree biomass modelling using airborne laser scanning. ISPRS J. Photogramm. Remote Sens. 2013, 85, 66–73. [Google Scholar] [CrossRef]
- Popescu, S.C.; Wynne, R.H.; Nelson, R.H. Measuring individual tree crown diameter with LiDAR and assessing its influence on estimating forest volume and biomass. Can. J. Remote Sens. 2003, 29, 564–577. [Google Scholar] [CrossRef]
- Yu, X.; Hyyppä, J.; Kaartinen, H.; Maltamo, M. Automatic detection of harvested trees and determination of forest growth using airborne laser scanning. Remote Sens. Environ. 2004, 90, 451–462. [Google Scholar] [CrossRef]
- Falkowski, M.; Hudak, A.; Crookston, N.; Gessler, P.; Smith, A. Landscape-scale parameterization of a tree-level forest growth model: A k-NN imputation approach incorporating LiDAR data. Can. J. For. Res. 2010, 40, 184–199. [Google Scholar] [CrossRef]
- Vepakomma, U.; St-Onge, B.; Kneeshaw, D. Response of a boreal forest to canopy gap openings: Assessing vertical and horizontal tree growth with multi-temporal LiDAR data. Ecolo. Appl. 2011, 21, 99–121. [Google Scholar] [CrossRef]
- Söderbergh, I.; Ledermann, T. Algorithms for simulating thinning and harvesting in five European individual-tree growth simulators: A review. Computers Electron. Agric. 2003, 39, 115–140. [Google Scholar] [CrossRef]
- Kärkkäinen, L.; Matala, J.; Härkönen, K.; Kellomäki, S.; Nuutinen, T. Potential recovery of industrial wood and energy wood raw material in different cutting and climate scenarios for Finland. Biomass Bioenergy 2008, 32, 934–943. [Google Scholar] [CrossRef]
- Lindberg, E.; Holmgren, J.; Olofsson, K.; Wallerman, J.; Olsson, H. Estimation of tree lists from airborne laser scanning by combining single-tree and area-based methods. Int. J. Remote Sens. 2010, 31, 1175–1192. [Google Scholar] [CrossRef]
- Næsset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Lim, K.; Treitz, P.; Wulder, M.; St-Onge, B.; Flood, M. LiDAR remote sensing of forest structure. Prog. Phys. Geogra. 2003, 27, 88–106. [Google Scholar] [CrossRef]
- Zimble, D.A.; Evans, D.L.; Carlson, G.C.; Parker, R.C.; Grado, S.C.; Gerard, P.D. Characterizing vertical forest structure using small-footprint airborne LiDAR. Remote Sens. Environ. 2003, 87, 171–182. [Google Scholar] [CrossRef]
- Coops, N.C.; Hilker, T.; Wulder, M.A.; St-Onge, B.; Newnham, G.; Siggins, A.; Trofymow, J.A. Estimating canopy structure of Douglas-fir forest stands from discrete-return LiDAR. Trees Struct. Funct. 2007, 21, 295–310. [Google Scholar] [CrossRef]
- Vastaranta, M.; Wulder, M.A.; White, J.; Pekkarinen, A.; Tuominen, S.; Ginzler, C.; Kankare, V.; Holopainen, M.; Hyyppä, J.; Hyyppä, H. Airborne laser scanning and digital stereo imagery measures of forest structure: Comparative results and implications to forest mapping and inventory update. Can. J. Remote Sens. 2013, 39, 382–395. [Google Scholar]
- White, J.C.; Wulder, M.A.; Vastaranta, M.; Coops, N.C.; Pitt, D.; Woods, M. The utility of image-based point clouds for forest inventory: A comparison with airborne laser scanning. Forests 2013, 3, 518–536. [Google Scholar]
- Wulder, M.A.; Coops, N.C.; Hudak, A.T.; Morsdorf, F.; Nelson, R.; Newnham, G.; Vastaranta, M. Status and prospects for LiDAR remote sensing of forested ecosystems. Can. J. Remote Sens. 2013, 39. [Google Scholar] [CrossRef]
- Jaskierniak, D.; Lane, P.N.J.; Robinson, A.; Lucieer, A. Extracting LiDAR indices to characterize multilayered forest structure using mixture distribution functions. Remote Sens. Environ. 2011, 115, 573–585. [Google Scholar] [CrossRef]
- Kraus, K.; Pfeifer, N. Determination of terrain models in wooded areas with airborne laser scanner data. ISPRS J. Photogramm. Remote Sens. 1998, 53, 193–203. [Google Scholar] [CrossRef]
- Lee, H.S.; Younan, N. DTM extraction of LiDAR returns via adaptive processing. IEEE Trans. Geosci. Remote Sens. 2003, 41, 2063–2069. [Google Scholar] [CrossRef]
- Chen, Q.; Gong, P.; Baldocchi, D.; Xie, G. Filtering airborne laser scanning data with morphological methods. Photogramm. Eng. Remote Sens. 2007, 73, 175–185. [Google Scholar] [CrossRef]
- Mongus, D.; Žalik, B. Parameter-free ground filtering of LiDAR data for automatic DTM generation. ISPRS J. Photogramm. Remote Sens. 2012, 67, 1–12. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.A.; Varhola, A.; Vastaranta, M.; Coops, N.C.; Cook, B.D.; Pitt, D.; Woods, M. A Best Practices Guide for Generating Forest Inventory Attributes from Airborne Laser Scanning Data Using the Area-Based Approach; Information Report FI-X-10; Natural Resources Canada, Canadian Forest Service, Canadian Wood Fibre Centre, Pacific Forestry Centre: Victoria, BC, Canada, 2013; p. 50. [Google Scholar]
- Hyyppä, J. Method for Determination of Stand Attributes and a Computer Program to Perform the Method. U.S. Patent 6,792,684, 28 October 1999. [Google Scholar]
- Mallet, C.; Bretar, F. Full-waveform topographic LiDAR: State-of-the-art. ISPRS J. Photogramm. Remote Sens. 2009, 64, 1–16. [Google Scholar] [CrossRef]
- Wagner, W.; Hollaus, M.; Briese, C.; Ducic, V. 3D vegetation mapping using small-footprint full-waveform airborne laser scanners. Int. J. Remote Sens. 2008, 29, 1433–1452. [Google Scholar] [CrossRef]
- Chauve, A.; Vega, C.; Durrieu, S.; Bretar, F.; Allouis, T.; Deseilligny, M.P.; Puech, W. Advanced full-waveform LiDAR data echo detection: Assessing quality of derived terrain and tree height models in an alpine coniferous forest. Int. J. Remote Sens. 2009, 30, 5211–5228. [Google Scholar] [CrossRef]
- Roncat, A.; Wagner, W.; Melzer, T.; Ullrich, A. Echo detection and localization in full-waveform airborne laser scanner data using the averaged square difference function estimator. Photogramm. J. Finl. 2008, 21, 62–75. [Google Scholar]
- Persson, A.; Söderman, U.; Töpel, J.; Ahlberg, S. Visualization and analysis of full-waveform airborne laser scanner data. In International Archives of Photogrammetry, Remote Sensing and Spatial Information Sciences; Copernicus Publications: Enschede, The Netherlands, 2005; Volume 36, (Part 3/W19); pp. 103–108. [Google Scholar]
- Reitberger, J.; Krzystek, P.; Stilla, U. Analysis of full waveform LiDAR data for tree species classification. In International Archives of Photogrammetry, Remote Sensing and Spatial Information Sciences; Copernicus Publications: Bonn, Germany, 2006; Volume 36, (Part 3); pp. 228–233. [Google Scholar]
- Reitberger, J.; Krzystek, P.; Stilla, U. Analysis of full waveform LiDAR data for the classification of deciduous and coniferous trees. Int. J. Remote Sens. 2008, 29, 1407–1431. [Google Scholar] [CrossRef]
- Rutzinger, M.; Höfle, B.; Hollaus, M.; Pfeifer, N. Object-based point cloud analysis of full-waveform airborne laser scanning data for urban vegetation classification. Sensors 2008, 8, 4505–4528. [Google Scholar] [CrossRef]
- Melzer, T. Non-parametric segmentation of ALS point clouds using mean shift. J. Appl. Geod. 2007, 1, 159–170. [Google Scholar]
- Lindberg, E.; Olofsson, K.; Holmgren, J.; Olsson, H. Estimation of 3D vegetation structure from waveform and discrete return airborne laser scanning data. Remote Sens. Environ. 2012, 118, 151–161. [Google Scholar] [CrossRef]
- Morsdorf, F.; Meier, E.; Kötz, B.; Itten, K.I.; Bobbertin, M.; Allgöwer, B. LiDAR-based geometric reconstruction of boreal type forest stands at single tree level for forest and wildland fire management. Remote Sens. Environ. 2004, 92, 353–362. [Google Scholar] [CrossRef]
- Reitberger, J.; Schnörr, Cl.; Krzystek, P.; Stilla, U. 3D segmentation of single trees exploiting full waveform LiDAR data. ISPRS J. Photogramm. Remote Sens. 2009, 64, 561–574. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, A. Identifying species of individual trees using airborne laser scanner. Remote Sens. Environ. 2004, 90, 415–423. [Google Scholar] [CrossRef]
- Brandtberg, T. Classifying individual tree species under leaf-off and leaf-on conditions using airborne LiDAR. ISPRS J. Photogramm. Remote Sens. 2007, 61, 325–340. [Google Scholar] [CrossRef]
- Ørka, H.O.; Naesset, E.; Bollandsas, O.M. Classifying species of individual trees by intensity and structure features derived from airborne laser scanner data. Remote Sens. Environ. 2009, 113, 1163–1174. [Google Scholar] [CrossRef]
- Kim, S.; McGaughey, R.J.; Andersen, H.E.; Schreuder, G. Tree species differentiation using intensity data derived from leaf-on and leaf-off airborne laser scanner data. Remote Sens. Environ. 2009, 113, 1575–1586. [Google Scholar] [CrossRef]
- Vaughn, N.R.; Moskal, L.M.; Turnblom, E.C. Tree species detection accuracies using discrete point LiDAR and airborne waveform LiDAR. Remote Sens. 2012, 4, 377–403. [Google Scholar] [CrossRef]
- Höfle, B.; Hollaus, M.; Lehner, H.; Pfeifer, N.; Wagner, W. Area-based parameterization of forest structure using full-waveform airborne laser scanning data. In Proceedings of SilviLaser 2008 the 8th International Conference on LiDAR Applications in Forest Assessment and Inventory, Edinburgh, Scotland, UK, 17–19 September 2008; pp. 229–235.
- Hollaus, M.; Mücke, W.; Höfle, B.; Dorigo, W.; Pfeifer, N.; Wagner, W.; Bauerhansl, C.; Regner, B. Tree species classification based on full-waveform airborne laser scanning data. In Proceedings of SilviLaser 2009the 9th International Conference on LiDAR Applications for Assessing Forest Ecosystems, College Station, TX, USA, 14–16 October 2009; pp. 54–62.
- Heinzel, J.; Koch, B. Exploring full-waveform LiDAR parameters for tree species classification. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 152–160. [Google Scholar] [CrossRef]
- Kaartinen, H.; Hyyppä, J. EuroSDR/ISPRS Project Commission II, Tree Extraction, Final Report. EuroSDR. 2008. Available online: http://bono.hostireland.com/~eurosdr/publications/53.pdf (accessed on 10 November 2012).
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Axelsson, P. DEM generation from laser scanner data using adaptive TIN models. In Proceedings of the XIXth ISPRS Congress Commission I–VII, Amsterdam, The Netherlands, 16–23 July 2000; pp. 110–117.
- Yu, X.; Hyyppä, J.; Holopainen, M.; Vastaranta, M.; Viitala, R. Predicting individual tree attributes from airborne laser point clouds based on random forest technique. ISPRS J. Photogramm. Remote Sens. 2011, 66, 28–37. [Google Scholar] [CrossRef]
- Yu, X.; Hyyppä, J.; Kukko, A.; Maltamo, M.; Kaartinen, H. Change detection techniques for canopy height growth measurements using airborne laser scanning data. Photogram. Eng. Remote Sens. 2006, 72, 1339–1348. [Google Scholar] [CrossRef]
- Chan, J.C.W.; Paelinckx, D. Evaluation of random forest and adaboost tree-based ensemble classification and spectral band selection for ecotope mapping using airborne hyperspectral imagery. Remote Sens. Environ. 2008, 112, 2999–3011. [Google Scholar] [CrossRef]
- Reese, H.; Nyström, M.; Nordkvist, K.; Olsson, H. Combining airborne laser scanning data and optical satellite data for classification of alpine vegetation. Int. J. Appl. Earth Obs. Geoinf. 2014, 27, 81–90. [Google Scholar] [CrossRef]
- Hudak, A.T.; Crookston, N.L.; Evans, J.S.; Hall, D.E.; Falkowski, M.J. Nearest neighbor imputation of species-level, plot-scale forest structure attributes from LiDAR data. Remote Sens. Environ. 2008, 112, 2232–2245. [Google Scholar] [CrossRef]
- Goutte, G.; Gaussier, E. A probabilistic interpretation of precision, recall and F-score, with implication for evaluation. Adv. Inf. Retr. 2005, 3408, 345–359. [Google Scholar] [CrossRef]
- Sokolova, M.; Japkowicz, N.; Szpakowicz, S. Beyond Accuracy, F-Score and ROC: A Family of Discriminant Measures for Performance Evaluation, AI 2006; Advances in Artificail Intelligence; Sattar, S., Kang, B.H., Eds.; Springer Berlin: Heidelberg, Germany, 2006; pp. 1015–1021. [Google Scholar]
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Kaartinen, H.; Hyyppä, J.; Yu, X.; Vastaranta, M.; Hyyppä, H.; Kukko, A.; Holopainen, M.; Heipke, C.; Hirschmugl, M.; Morsdorf, F.; et al. An international comparison of individual tree detection and extraction using airborne laser scanning. Remote Sens. 2012, 4, 950–974. [Google Scholar] [CrossRef] [Green Version]
- Heinzel, J.N.; Weinacker, H.; Koch, B. Prior-knowledge-based single-tree extraction. Int. J. Remote Sens. 2011, 32, 4999–5020. [Google Scholar] [CrossRef]
- Falkowski, M.J.; Smith, A.M.S.; Gessler, P.E.; Hudak, A.T.; Vierling, L.A.; Evans, J.S. The influence of conifer forest canopy cover on the accuracy of two individual tree measurement algorithms using LiDAR data. Can. J. Remote Sens. 2008, 34, 338–350. [Google Scholar] [CrossRef]
- Höfle, B.; Hollaus, M.; Hagenauer, J. Urban vegetation detection using radiometrically calibrated smallfootprint full-waveform airborne LiDAR data. ISPRS J. Photogramm. Remote Sens. 2012, 67, 134–147. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yu, X.; Litkey, P.; Hyyppä, J.; Holopainen, M.; Vastaranta, M. Assessment of Low Density Full-Waveform Airborne Laser Scanning for Individual Tree Detection and Tree Species Classification. Forests 2014, 5, 1011-1031. https://doi.org/10.3390/f5051011
Yu X, Litkey P, Hyyppä J, Holopainen M, Vastaranta M. Assessment of Low Density Full-Waveform Airborne Laser Scanning for Individual Tree Detection and Tree Species Classification. Forests. 2014; 5(5):1011-1031. https://doi.org/10.3390/f5051011
Chicago/Turabian StyleYu, Xiaowei, Paula Litkey, Juha Hyyppä, Markus Holopainen, and Mikko Vastaranta. 2014. "Assessment of Low Density Full-Waveform Airborne Laser Scanning for Individual Tree Detection and Tree Species Classification" Forests 5, no. 5: 1011-1031. https://doi.org/10.3390/f5051011






