Transcriptome Sequencing and Comparative Analysis of Piptoporus betulinus in Response to Birch Sawdust Induction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strain
2.2. RNA Extraction
2.3. Transcriptome Analysis
2.4. Quantitative Real-Time PCR Validation
2.5. Availability of Data
3. Results
3.1. Raw Reads Processing and de novo Assembly
3.2. Functional Annotation
3.3. Changes in Gene Expression under Sawdust Induction
3.4. Detection of Lignocellulose-Degrading Enzyme-Relevant Gene Sequences and qRT-PCR Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lemieszek, M.; Langner, E.; Kaczor, J.; Kandefer-Szerszen, M.; Sanecka, B.; Mazurkiewicz, W.; Rzeski, W. Anticancer effect of fraction isolated from medicinal birch polypore mushroom, Piptoporus betulinus (bull.: Fr.) p. Karst. (aphyllophoromycetideae): In Vitro studies. Int. J. Med. Mushrooms 2009, 11, 351–364. [Google Scholar] [CrossRef]
- Wangun, H.V.K.; Berg, A.; Hertel, W.; Nkengfack, A.E.; Hertweck, C. Anti-inflammatory and anti-hyaluronate lyase activities of lanostanoids from Piptoporus betulinus. J. Antibiot. 2004, 57, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Liu, X.; Wang, Q. Identification of wood decay related genes from Piptoporus betulinus (bull. Fr.) karsten using differential display reverse transcription pcr (ddrt-pcr). Biotechnol. Biotechnol. Equip. 2012, 26, 2961–2965. [Google Scholar] [CrossRef]
- Shang, J.; Yan, S.; Wang, Q. Degradation mechanism and chemical component changes in Betula platyphylla wood by wood-rot fungi. BioResources 2013, 8, 6066–6077. [Google Scholar] [CrossRef]
- Wymelenberg, A.V.; Gaskell, J.; Mozuch, M.; Bondurant, S.S.; Sabat, G.; Ralph, J.; Skyba, O.; Mansfield, S.D.; Blanchette, R.A.; Grigoriev, I.V.; Kersten, P.J.; Cullen, D. Significant alteration of gene expression in wood decay fungi Postia placenta and Phanerochaete chrysosporium by plant species. Appl. Environ. Microbiol. 2011, 77, 4499–4507. [Google Scholar] [CrossRef] [PubMed]
- Klinke, H.B.; Thomsen, A.B.; Ahring, B.K. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl. Microbiol. Biotechnol. 2004, 66, 10–26. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Kanawaku, R.; Masumoto, M.; Yanase, H. Efficient xylose fermentation by the brown rot fungus Neolentinus lepideus. Enzym. Microb. Technol. 2012, 50, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Dashtban, M.; Schraft, H.; Qin, W. Fungal bioconversion of lignocellulosic residues; opportunities & perspectives. Int. J. Biol. Sci. 2009, 5, 578–595. [Google Scholar] [PubMed]
- Canam, T.; Town, J.; Iroba, K.; Tabil, L.; Dumonceaux, T. Pretreatment of lignocellulosic biomass using microorganisms: Approaches, advantages, and limitations. Sustain. Degrad. Lignocellul. Biomass-Techn. Appl. Commer. 2013. [Google Scholar] [CrossRef]
- Zeng, X.X.; Tang, J.X.; Yin, H.Q.; Liu, X.D.; Pei, J.; Liu, H.W. Isolation, identification and cadmium adsorption of a high cadmium-resistant Paecilomyces lilacinus. Afr. J. Biotechnol. 2010, 9, 6525–6533. [Google Scholar]
- Kim, S.Y.; Park, S.Y.; Ko, K.S.; Jung, H.S. Phylogenetic analysis of antrodia and related taxa based on partial mitochondrial ssu rDNA sequences. Antonie Leeuwenhoek 2003, 83, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Korripally, P.; Hunt, C.G.; Houtman, C.J.; Jones, D.C.; Kitin, P.J.; Cullen, D.; Hammel, K.E. Regulation of gene expression during the onset of ligninolytic oxidation by Phanerochaete chrysosporium on spruce wood. Appl. Environ. Microbiol. 2015, 81, 7802–7812. [Google Scholar] [CrossRef] [PubMed]
- Kuuskeri, J.; Häkkinen, M.; Laine, P.; Smolander, O.P.; Tamene, F.; Miettinen, S.; Nousiainen, P.; Kemell, M.; Auvinen, P.; Lundell, T. Time-scale dynamics of proteome and transcriptome of the white-rot fungus Phlebia radiata: Growth on spruce wood and decay effect on lignocellulose. Biotechnol. Biofuels 2016, 9, 192. [Google Scholar] [CrossRef] [PubMed]
- Gaskell, J.; Blanchette, R.A.; Stewart, P.E.; Bondurant, S.; Adams, M.; Sabat, G.; Kersten, P.; Cullen, D. Transcriptome and secretome analyses of the wood decay fungus Wolfiporia cocos support alternative mechanisms of lignocellulose conversion. Appl. Environ. Microbiol. 2016, 82, 3979–3987. [Google Scholar] [CrossRef] [PubMed]
- Skyba, O.; Cullen, D.; Douglas, C.J.; Mansfield, D. Gene expression patterns of wood decay fungi Postia placenta and Phanerochaete chrysosporium are influenced by wood substrate composition during degradation. Appl. Environ. Microbiol. 2016, 82, 4387–4400. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhu, H.; Ruan, J.; Qian, W.; Fang, X.; Shi, Z.; Li, Y.; Li, S.; Shan, G.; Kristiansen, K. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010, 20, 265–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B. Tigr gene indices clustering tools (TGICL): A software system for fast clustering of large est datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2Go: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L. Wego: A web tool for plotting go annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with tophat and cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Yao, J.; Wang, X.; Guo, H.; Duan, D. Transcriptome sequencing and comparative analysis of Saccharina japonica (laminariales, phaeophyceae) under blue light induction. PLoS ONE 2012, 7, e39704. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔδCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Cong, B.; Wang, N.; Liu, S.; Liu, F.; Yin, X.; Shen, J. Isolation, characterization and transcriptome analysis of a novel Antarctic Aspergillus sydowii strain MS-19 as a potential lignocellulosic enzyme source. BMC Microbiol. 2017, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.; Knackmuss, H.J. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem. J. 1980, 192, 339. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, S.C.; Coemans, B.; Podevin, N.; Laukens, K.; Witters, E.; Matsumura, H. Functional genomics in a non-model crop: Transcriptomics or proteomics? Physiol. Plant. 2008, 133, 117. [Google Scholar] [CrossRef] [PubMed]
- Hori, C.; Gaskell, J.; Igarashi, K.; Samejima, M.; Hibbett, D.; Henrissat, B.; Cullen, D. Genomewide analysis of polysaccharides degrading enzymes in 11 white-and brown-rot Polyporales provides insight into mechanisms of wood decay. Mycologia 2013, 105, 1412–1427. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.; Challacombe, J.; Morgenstern, I.; Hibbett, D.; Schmoll, M.; Kubicek, C.P.; Ferreira, P.; Ruiz-Duenas, F.J.; Martinez, A.T.; Kersten, P. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc. Natl. Acad. Sci. USA 2009, 106, 1954–1959. [Google Scholar] [CrossRef] [PubMed]
- Rytioja, J.; Hildén, K.; Yuzon, J.; Hatakka, A.; de Vries, R.P.; Mäkelä, M.R. Plant-polysaccharide-degrading enzymes from basidiomycetes. Microbiol. Mol. Biol. Rev. 2014, 78, 614–649. [Google Scholar] [CrossRef] [PubMed]
- Yelle, D.J.; Wei, D.; John, R.; Hammel, K.E. Multidimensional nmr analysis reveals truncated lignin structures in wood decayed by the brown rot basidiomycete Postia placenta. Environ. Microbiol. 2011, 13, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Presley, G.N.; Hammel, K.E.; Ryu, J.S.; Menke, J.R.; Figueroa, M.; Hu, D.; Orr, G.; Schilling, J.S. Localizing gene regulation reveals a staggered wood decay mechanism for the brown rot fungus Postia placenta. Proc. Natl. Acad. Sci. USA 2016, 113, 10968. [Google Scholar] [CrossRef] [PubMed]
- Tuor, U.; Winterhalter, K.; Fiechter, A. Enzymes of white-rot fungi involved in lignin degradation and ecological determinants for wood decay. J. Biotechnol. 1995, 41, 1–17. [Google Scholar] [CrossRef]
- Lee, D.; Meyer, K.; Chapple, C.; Douglas, C.J. Antisense suppression of 4-coumarate:Coenzyme a ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 1997, 9, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- An, H.D.; Wei, D.S.; Xiao, T.T. Transcriptional profiles of laccase genes in the brown rot fungus Postia placenta mad-r-698. J. Microbiol. 2015, 53, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Prince, R.C. Bioremediation: An overview of how microbiological processes can be applied to the cleanup of organic and inorganic environmental pollutants. In Encyclopedia of Environmental Microbiology; John Wiley & Sons, lnc.: Hoboken, NJ, USA, 2003. [Google Scholar]
- Wang, C.; Jin-Ying, X.I.; Hong-Ying, H.U.; Wen, X.H. Biodegradation of gaseous chlorobenzene by white-rot fungus Phanerochaete chrysosporium. Biomed. Environ. Sci. 2008, 21, 474–478. [Google Scholar] [CrossRef]
- Hadibarata, T.; Teh, Z.C.; Rubiyatno; Zubir, M.M.; Khudhair, A.B.; Yusoff, A.R.; Salim, M.R.; Hidayat, T. Identification of naphthalene metabolism by white rot fungus Pleurotus eryngii. Bioprocess Biosyst. Eng. 2013, 36, 1455–1461. [Google Scholar] [CrossRef] [PubMed]
- Hadibarata, T.; Yusoff, A.R.M.; Aris, A.; Kristanti, R.A. Identification of naphthalene metabolism by white rot fungus Armillaria sp.F022. J. Environ. Sci. 2012, 24, 728–732. [Google Scholar] [CrossRef]
- Krueger, M.C.; Hofmann, U.; Moeder, M.; Schlosser, D. Potential of wood-rotting fungi to attack polystyrene sulfonate and its depolymerisation by Gloeophyllum trabeum via hydroquinone-driven fenton chemistry. PLoS ONE 2015, 10, e0131773. [Google Scholar] [CrossRef] [PubMed]
- Ide, M.; Ichinose, H.; Wariishi, H. Molecular identification and functional characterization of cytochrome P450 monooxygenases from the brown-rot basidiomycete Postia placenta. Arch. Microbiol. 2012, 194, 243–253. [Google Scholar] [CrossRef] [PubMed]
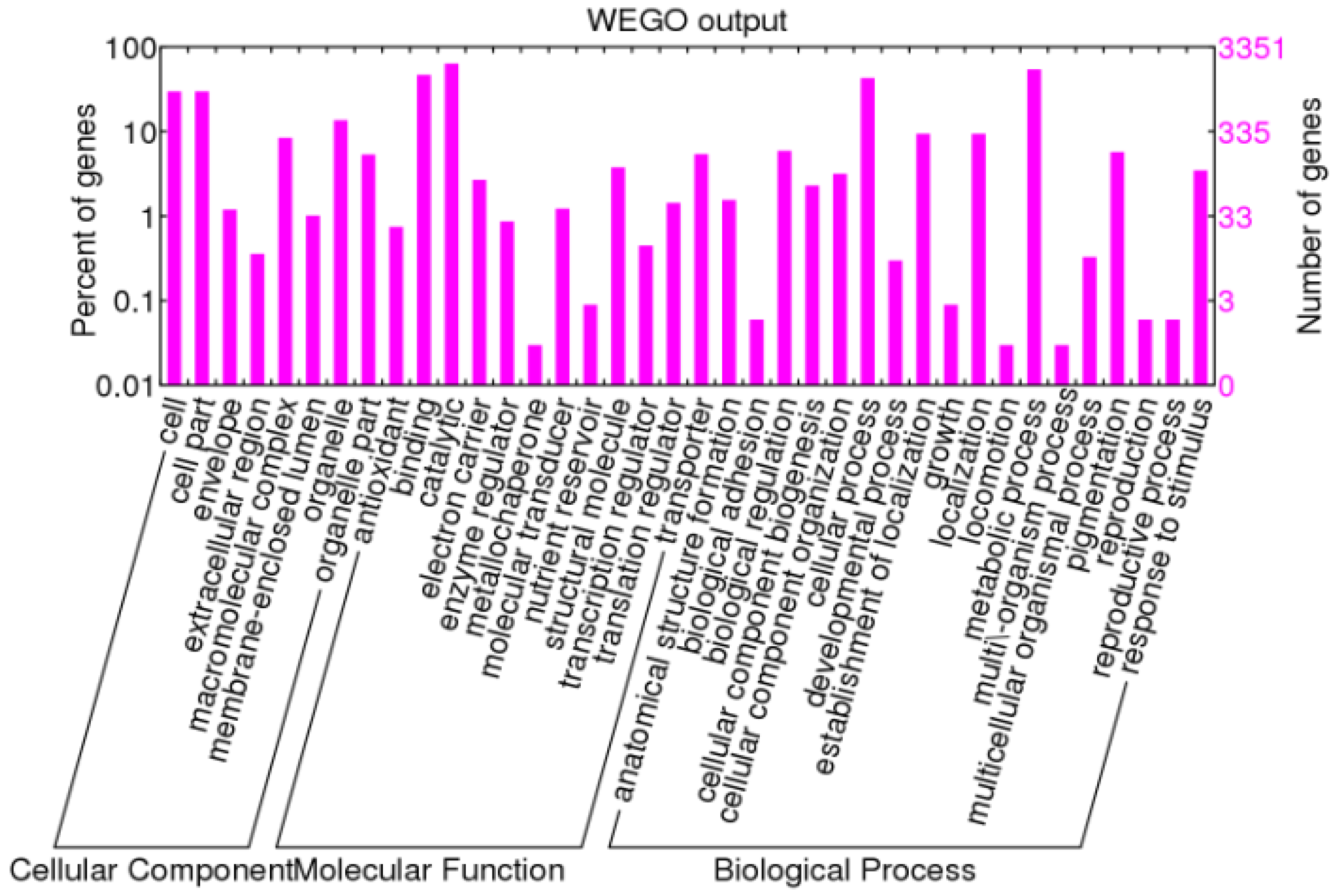
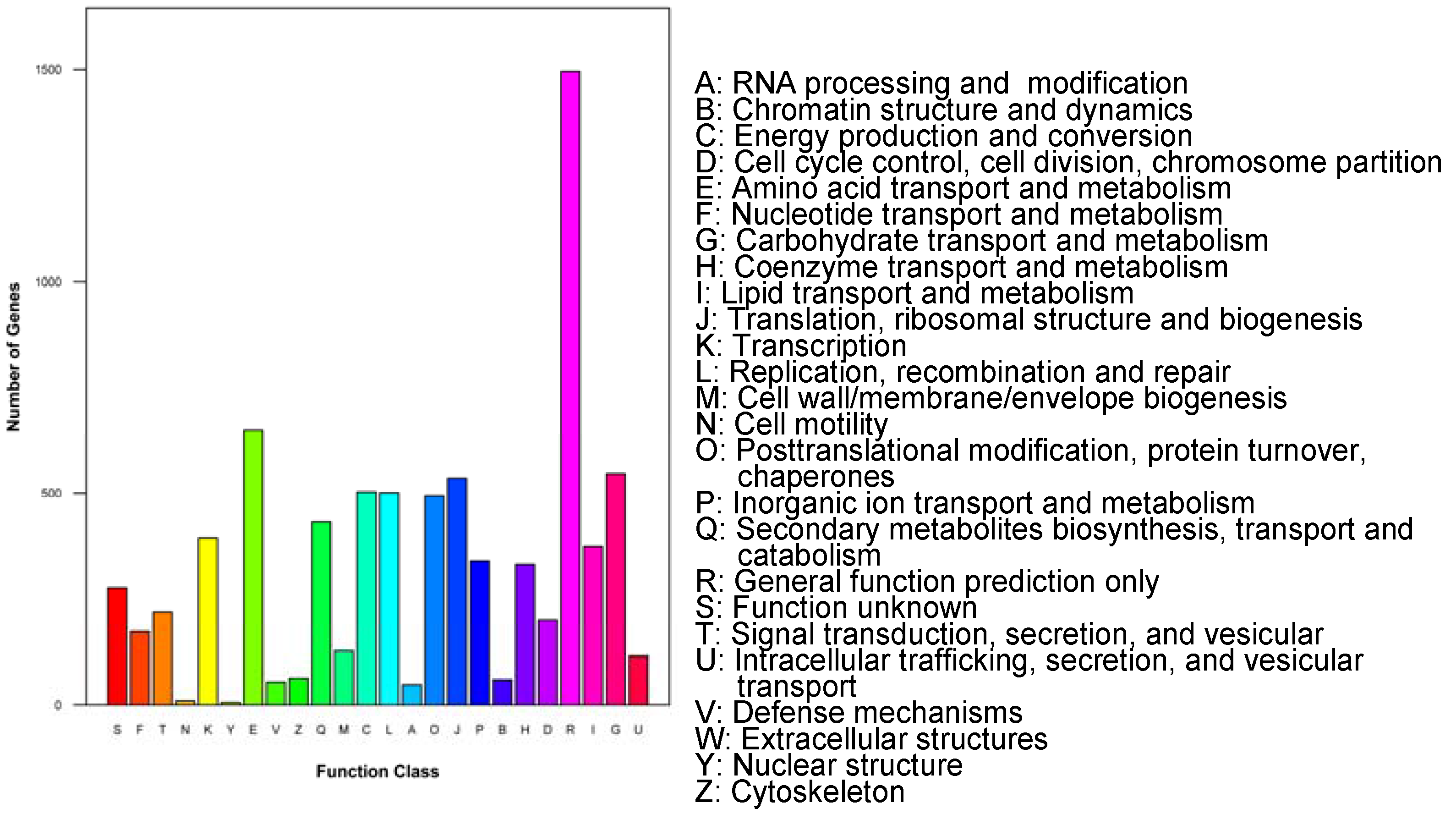
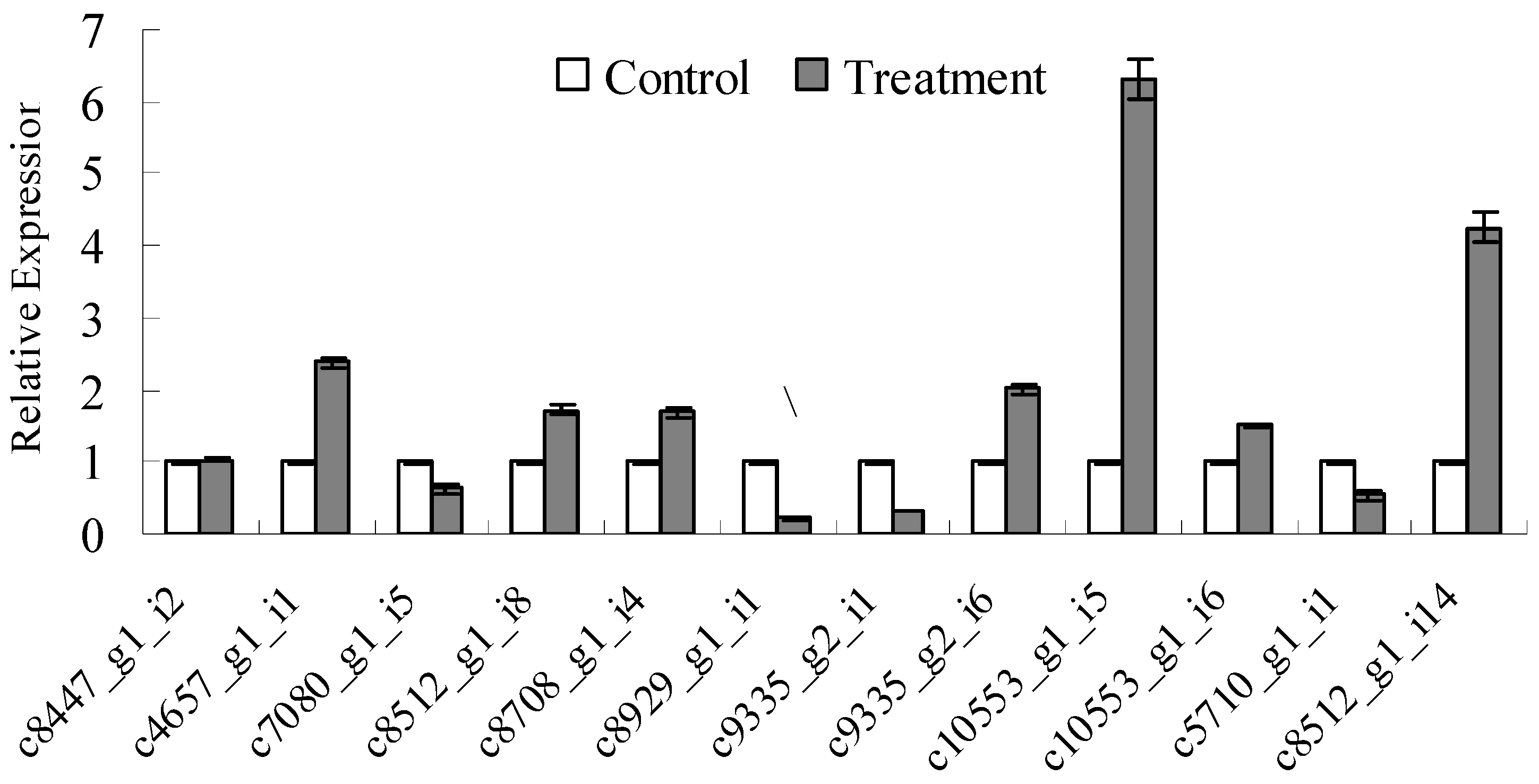
| Length (bp) | Total Number | Percentage (%) |
|---|---|---|
| <200 | 0 | 0.0 |
| 200–500 | 4800 | 21.9 |
| 500–1000 | 3362 | 15.4 |
| 1000–1500 | 3049 | 23.9 |
| 1500–2000 | 2626 | 12.0 |
| ≥2000 | 8044 | 36.8 |
| Total | 21,882 | |
| Total length of all unigenes | 42,652,276 | |
| Median length of all unigenes (N50) | 3007 | |
| Average length of all unigenes | 1949.17 | |
| Database | Number of Annotated Unigenes | Percentage of Annotated Unigenes |
|---|---|---|
| Nr | 21,255 | 97.1% |
| Swissport | 3351 | 15.3% |
| KEGG | 6256 | 28.6% |
| COG | 7955 | 36.4% |
| Enzymes | Number of Unigenes |
|---|---|
| glycoside hydrolases | 109 |
| glycosyl transferases | 37 |
| carbohydrate esterases | 12 |
| polysaccharide lyases | 13 |
| beta-glucosidase | 52 |
| alpha-glucosidase | 1 |
| mannosidase | 16 |
| glycosyltransferase | 4 |
| polygalacturonase | 11 |
| alpha-galactosidase | 7 |
| D-xylose 1-dehydrogenase (NADP) | 4 |
| N-glycosylase | 2 |
| Pectinesterase | 9 |
| 3-(3-hydroxy-phenyl)propionate hydroxylase | 22 |
| alpha-amylase | 5 |
| xylose isomerase | 7 |
| beta-xylosidase | 2 |
| xyloglucan:xyloglucosyl transferase | 2 |
| glucosylceramidase | 5 |
| chitinase | 20 |
| Total | 340 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Peng, M.; Shah, S.S.; Wang, Q. Transcriptome Sequencing and Comparative Analysis of Piptoporus betulinus in Response to Birch Sawdust Induction. Forests 2017, 8, 374. https://doi.org/10.3390/f8100374
Yang L, Peng M, Shah SS, Wang Q. Transcriptome Sequencing and Comparative Analysis of Piptoporus betulinus in Response to Birch Sawdust Induction. Forests. 2017; 8(10):374. https://doi.org/10.3390/f8100374
Chicago/Turabian StyleYang, Lixia, Mu Peng, Syed Sadaqat Shah, and Qiuyu Wang. 2017. "Transcriptome Sequencing and Comparative Analysis of Piptoporus betulinus in Response to Birch Sawdust Induction" Forests 8, no. 10: 374. https://doi.org/10.3390/f8100374





