Environmental Factors Driving the Recovery of Bay Laurels from Phytophthora ramorum Infections: An Application of Numerical Ecology to Citizen Science
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samplings and Laboratory Analyses
2.2. Design of the Algoritm to Detect and Geolocate the Recovered Trees
2.2.1. Rationale
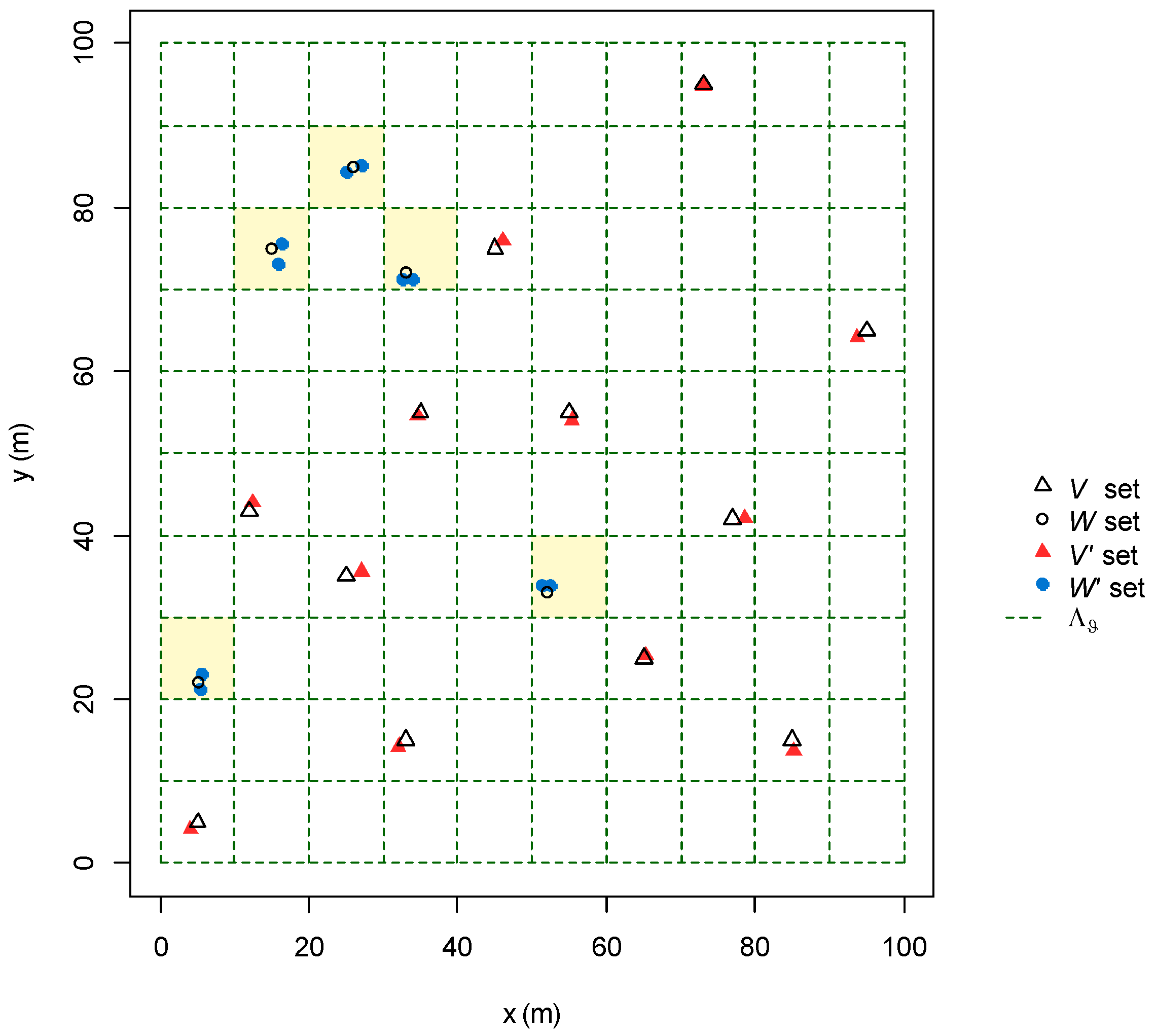
2.2.2. Modelling GPS Error and Spatial Resolution of the Grid-System
2.2.3. Design of the Algorithm
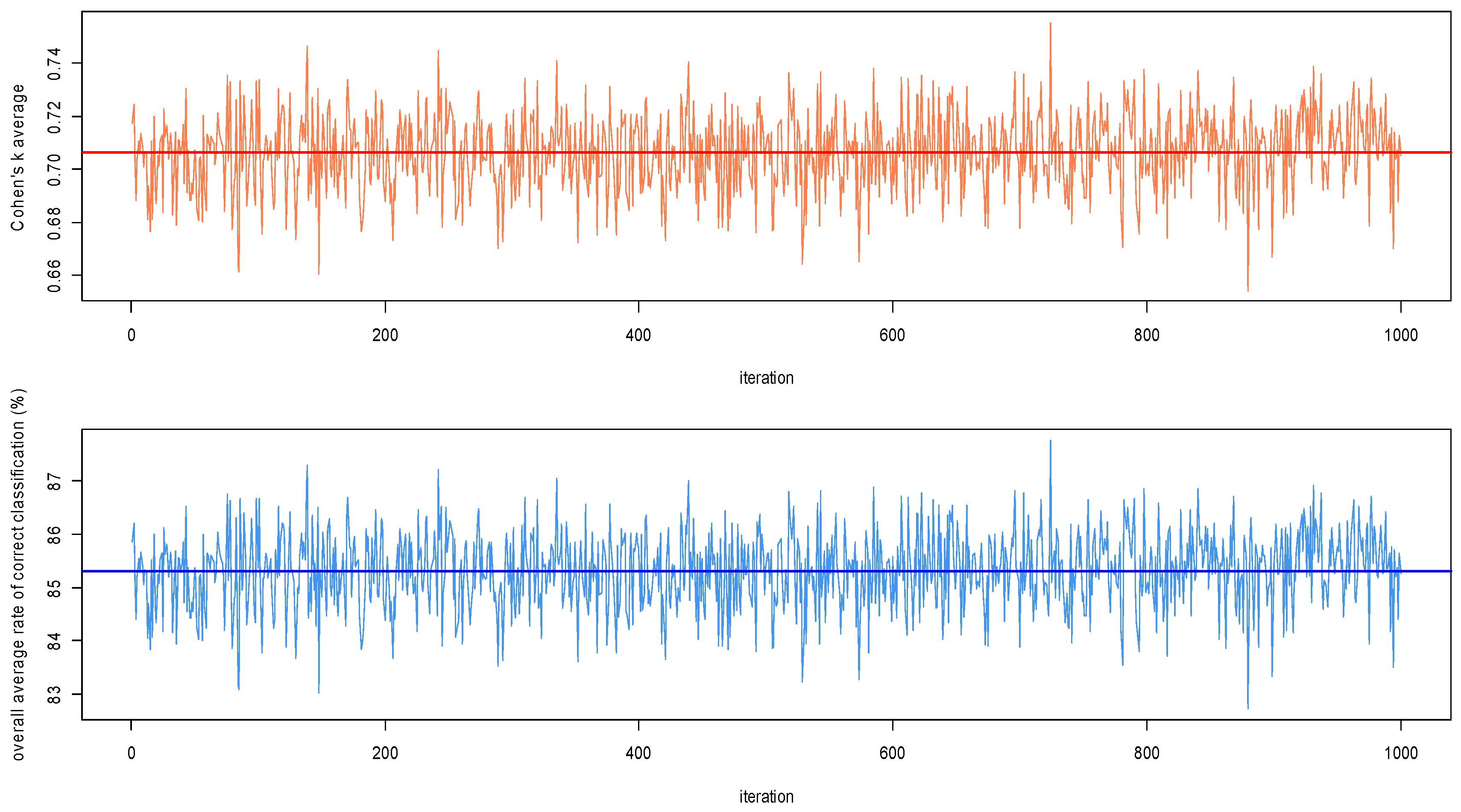
2.2.4. Algorithm Performances Assessment
2.3. Detection and Geolocation of the Trees Recovered from P. ramorum Infections Based on the “SODmap” Database
2.4. Analysis of the Association between Environmental Factors and Recovery from P. ramorum Infections
2.5. Software and Libraries Used for Statistical, GIS and Numerical Analyses
3. Results
3.1. Design of the Algorithm to Detect and Geolocate the Recovered Trees
3.1.1. GPS Error and Spatial Resolution of the Grid-System
3.1.2. Design of the Algorithm
3.1.3. Algorithm Performances Assessment
3.2. Detection and Geolocation of Trees Recovered from P. ramorum Infections Based on the “SODmap” Database
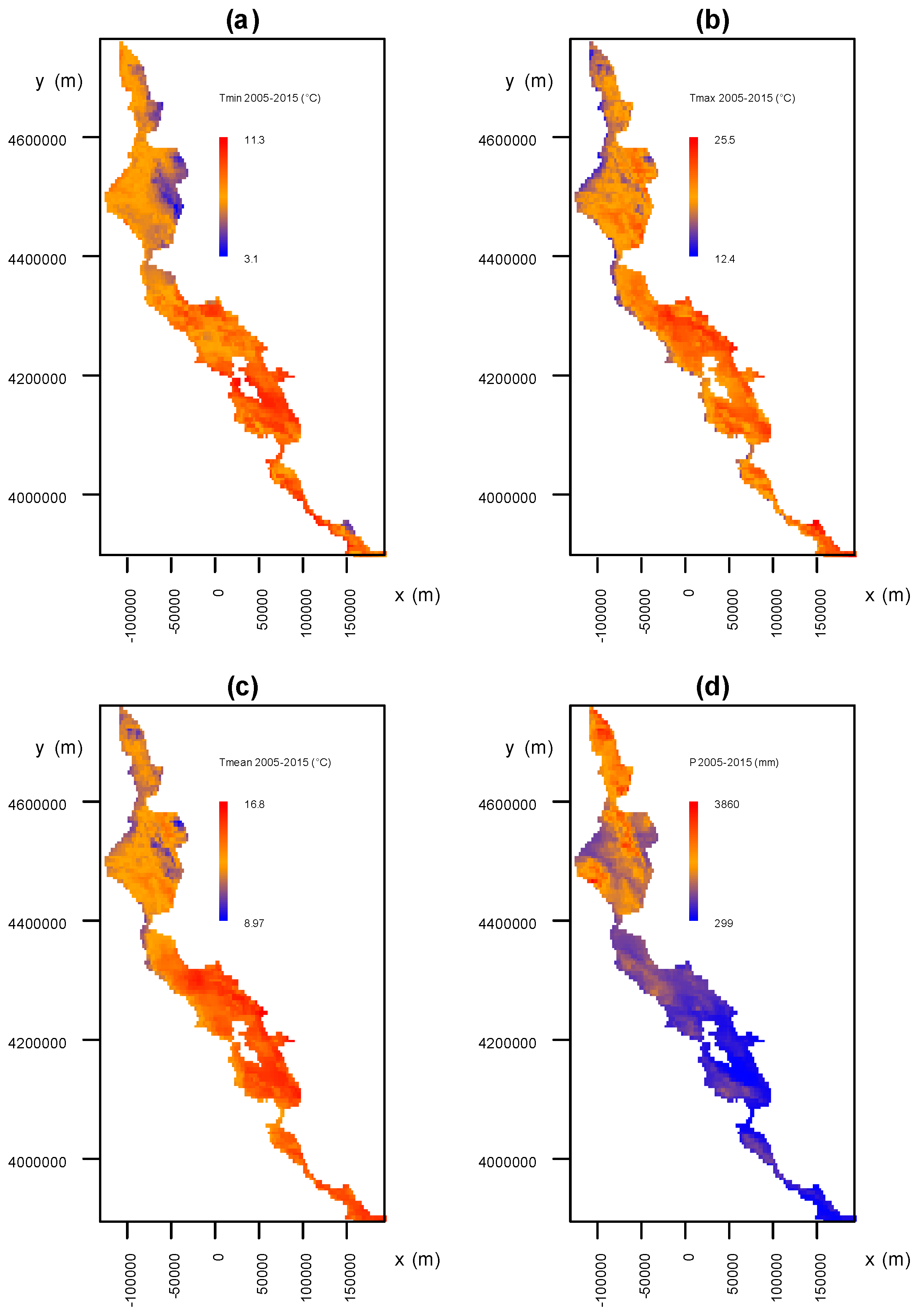
3.3. Analysis of the Association between Environmental Factors and Recovery from P. ramorum Infections
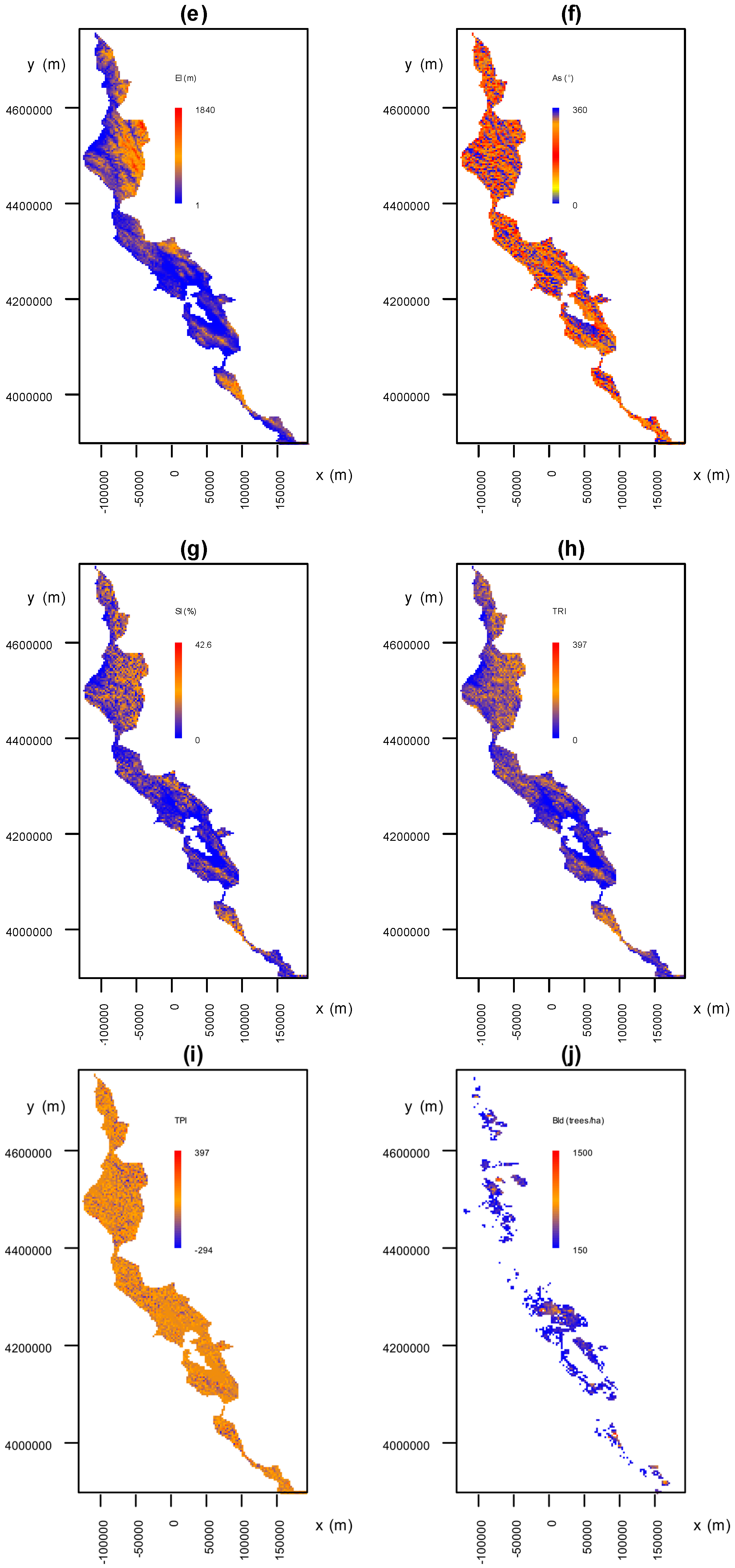
3.3.1. Environmental Factors
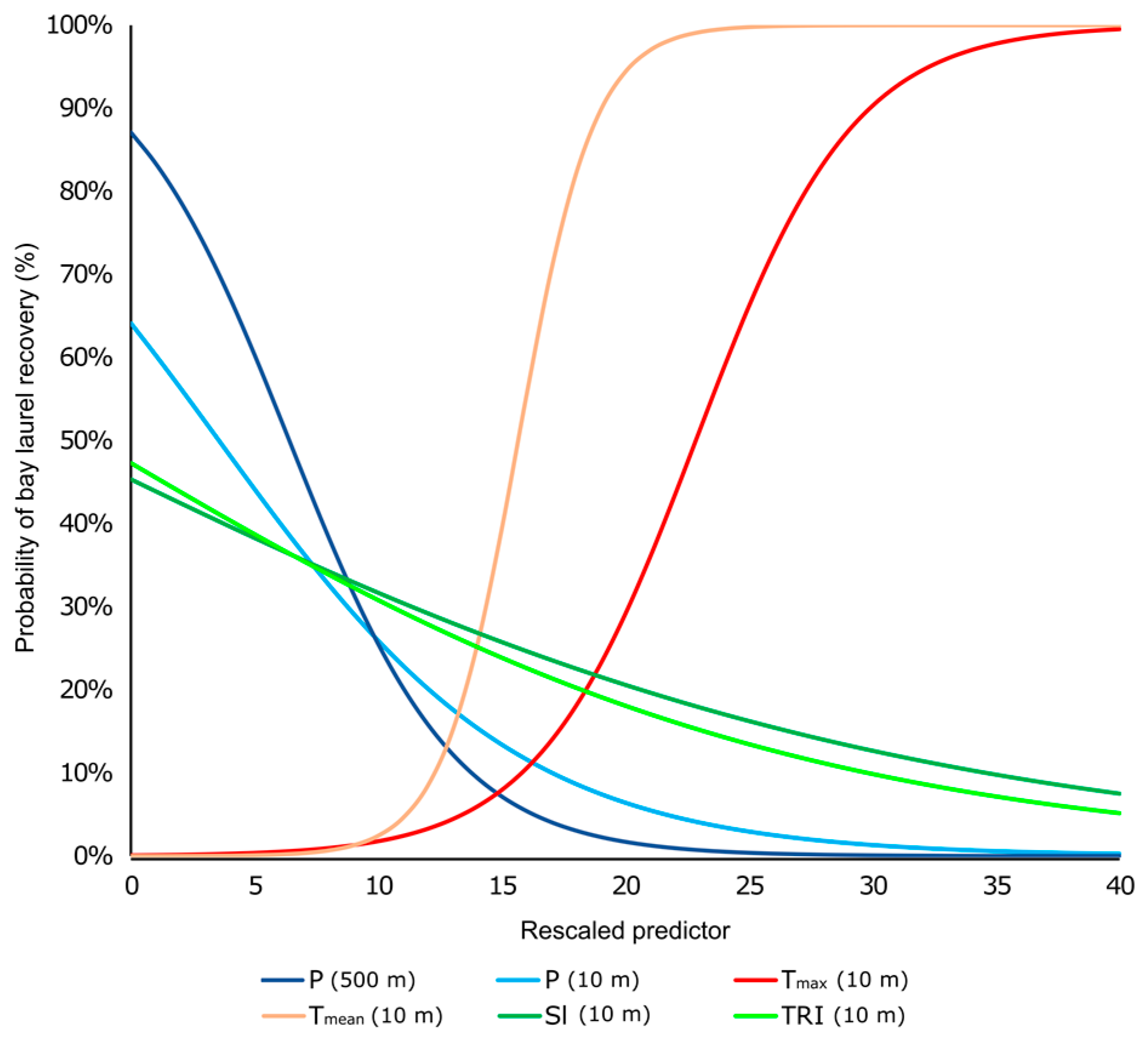
3.3.2. Binary Logistic Regression Models
3.3.3. Aspect Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- McNew, G.L. The nature, origin and evolution of parasitism. In Plant Pathology: An Advanced Treatise; Horsfall, J.G., Dimond, A.E., Eds.; Academic Press: New York, NY, USA, 1960; pp. 19–69. [Google Scholar]
- Scholthof, K.G. The disease triangle: Pathogens, the environment and society. Nat. Rev. Microbiol. 2007, 5, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, D.M.; Garbelotto, M.; Hansen, E.M. Phytophthora ramorum: Integrative research and management of an emerging pathogen in California and Oregon forests. Annu. Rev. Phytopathol. 2005, 43, 309–335. [Google Scholar] [CrossRef] [PubMed]
- Gonthier, P.; Nicolotti, G.; Linzer, R.; Guglielmo, F.; Garbelotto, M. Invasion of European pine stands by a North American forest pathogen and its hybridization with a native interfertile taxon. Mol. Ecol. 2007, 16, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, T.; Holdenrieder, O. Pathogenicity of Chalara fraxinea. For. Pathol. 2009, 39, 1–7. [Google Scholar] [CrossRef]
- Visentin, I.; Gentile, S.; Valentino, D.; Gonthier, P.; Tamietti, G.; Cardinale, F. Gnomoniopsis castanea sp. nov. (Gnomoniaceae, Diaporthales) as the causal agent of nut rot in sweet chestnut. J. Plant Pathol. 2012, 94, 411–419. [Google Scholar]
- Hauptman, T.; Piškur, B.; Groot, M.D.; Ogris, N.; Ferlan, M.; Jurc, D. Temperature effect on Chalara fraxinea: Heat treatment of saplings as a possible disease control method. For. Pathol. 2013, 43, 360–370. [Google Scholar]
- Santini, A.; Ghelardini, L.; Pace, C.D.; Desprez-Loustau, M.L.; Capretti, P.; Chandelier, A.; Cech, T.; Chira, D.; Diamandis, S.; Gaitniekis, T.; et al. Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytol. 2012, 197, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Gonthier, P.; Anselmi, N.; Capretti, P.; Bussotti, F.; Feducci, M.; Giordano, L.; Honorati, T.; Lione, G.; Luchi, N.; Michelozzi, M.; et al. An integrated approach to control the introduced forest pathogen Heterobasidion irregulare in Europe. Forestry 2014, 87, 471–481. [Google Scholar] [CrossRef]
- Garbelotto, M.; Guglielmo, F.; Mascheretti, S.; Croucher, P.J.P.; Gonthier, P. Population genetic analyses provide insights on the introduction pathway and spread patterns of the North American forest pathogen Heterobasidion irregulare in Italy. Mol. Ecol. 2013, 22, 4855–4869. [Google Scholar] [CrossRef] [PubMed]
- Lione, G.; Giordano, L.; Sillo, F.; Gonthier, P. Testing and modelling the effects of climate on the incidence of the emergent nut rot agent of chestnut Gnomoniopsis castanea. Plant Pathol. 2015, 64, 852–863. [Google Scholar] [CrossRef]
- Sillo, F.; Giordano, L.; Zampieri, E.; Lione, G.; De Cesare, S.; Gonthier, P. HRM analysis provides insights on the reproduction mode and the population structure of Gnomoniopsis castaneae in Europe. Plant Pathol. 2016, 66, 293–303. [Google Scholar] [CrossRef]
- Legendre, P.; Legendre, L. Numerical Ecology; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Kéry, M. Introduction to WinBUGS for Ecologists: Bayesian Approach to Regression, ANOVA, Mixed Models and Related Analyses; Academic Press: London, UK, 2010. [Google Scholar]
- Gonthier, P.; Brun, F.; Lione, G.; Nicolotti, G. Modelling the incidence of Heterobasidion annosum butt rots and related economic losses in alpine mixed naturally regenerated forests of northern Italy. For. Pathol. 2012, 42, 57–68. [Google Scholar] [CrossRef]
- Gonthier, P.; Lione, G.; Giordano, L.; Garbelotto, M. The American forest pathogen Heterobasidion irregulare colonizes unexpected habitats after its introduction in Italy. Ecol. Appl. 2012, 22, 2135–2143. [Google Scholar] [CrossRef] [PubMed]
- Meentemeyer, R.K.; Dorning, M.A.; Vogler, J.B.; Schmidt, D.; Garbelotto, M. Citizen science helps predict risk of emerging infectious disease. Front. Ecol. Environ. 2015, 13, 189–194. [Google Scholar] [CrossRef]
- Lione, G.; Gonthier, P. A permutation-randomization approach to test the spatial distribution of plant diseases. Phytopathology 2016, 106, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Lione, G.; Giordano, L.; Ferracini, C.; Alma, A.; Gonthier, P. Testing ecological interactions between Gnomoniopsis castaneae and Dryocosmus kuriphilus. Acta Oecol. 2016, 77, 10–17. [Google Scholar] [CrossRef]
- Paparella, F.; Ferracini, C.; Portaluri, A.; Manzo, A.; Alma, A. Biological control of the chestnut gall wasp with T. sinensis: A mathematical model. Ecol. Model. 2016, 338, 17–36. [Google Scholar] [CrossRef]
- Zampieri, E.; Giordano, L.; Lione, G.; Vizzini, A.; Sillo, F.; Balestrini, R.; Gonthier, P. A nonnative and a native fungal plant pathogen similarly stimulate ectomycorrhizal development but are perceived differently by a fungal symbiont. New Phytol. 2017, 213, 1836–1849. [Google Scholar] [CrossRef] [PubMed]
- Garbelotto, M.; Schmidt, D.; Swain, S.; Hayden, K.; Lione, G. The ecology of infection between a transmissive and a dead-end host provides clues for the treatment of a plant disease. Ecosphere 2017, 8, e01815. [Google Scholar] [CrossRef]
- Benda, G.T.A.; Naylor, A.W. On the tobacco ringspot disease. III. Heat and recovery. Am. J. Bot. 1958, 45, 33–37. [Google Scholar] [CrossRef]
- Ghoshal, B.; Sanfaçon, H. Temperature-dependent symptom recovery in Nicotiana benthamiana plants infected with tomato ringspot virus is associated with reduced translation of viral RNA2 and requires ARGONAUTE 1. Virology 2014, 456, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Chinnaraja, C.; Viswanathan, R. Variability in yellow leaf symptom expression caused by the sugarcane yellow leaf virus and its seasonal influence in sugarcane. Phytoparasitica 2015, 43, 339–353. [Google Scholar] [CrossRef]
- Morone, C.; Boveri, M.; Giosuè, S.; Gotta, P.; Rossi, V.; Scapin, I.; Marzachì, C. Epidemiology of flavescence dorée in vineyards in northwestern Italy. Phytopathology 2007, 97, 1422–1427. [Google Scholar] [CrossRef] [PubMed]
- Galetto, L.; Miliordos, D.; Roggia, C.; Rashidi, M.; Sacco, D.; Marzachì, C.; Bosco, D. Acquisition capability of the grapevine Flavescence dorée by the leafhopper vector Scaphoideus titanus Ball correlates with phytoplasma titre in the source plant. J. Pest Sci. 2014, 87, 671–679. [Google Scholar] [CrossRef]
- Karajeh, M.R.; Al-Momany, A.R. Effect of postplanting soil solarization and solar chamber on Verticillium wilt of olive. Jordan J. Agric. Sci. 2008, 4, 335–342. [Google Scholar]
- Aguayo, J.; Elegbede, F.; Husson, C.; Saintonge, F.X.; Marçais, B. Modeling climate impact on an emerging disease, the Phytophthora alni-induced alder decline. Glob. Chang. Biol. 2014, 20, 3209–3221. [Google Scholar] [CrossRef] [PubMed]
- Rosenvald, R.; Drenkhan, R.; Riit, T.; Lõhmusc, A. Towards silvicultural mitigation of the European ash (Fraxinus excelsior) dieback: The importance of acclimated trees in retention forestry. Can. J. For. Res. 2015, 45, 1206–1214. [Google Scholar] [CrossRef]
- Brown, N.; Jeger, M.; Kirk, S.; Xu, X.; Denman, S. Spatial and temporal patterns in symptom expression within eight woodlands affected by Acute Oak Decline. For. Ecol. Manag. 2016, 360, 97–109. [Google Scholar] [CrossRef]
- Venette, R.C.; Cohen, S.D. Potential climatic suitability for establishment of Phytophthora ramorum within the contiguous United States. For. Ecol. Manag. 2006, 231, 18–26. [Google Scholar] [CrossRef]
- Rizzo, D.M.; Garbelotto, M.; Davidson, J.M.; Slaughter, G.W.; Koike, S.T. Phytophthora ramorum as the cause of extensive mortality of Quercus spp. and Lithocarpus densiflorus in California. Plant Dis. 2002, 86, 205–214. [Google Scholar] [CrossRef]
- Rizzo, D.M.; Garbelotto, M.; Davidson, J.M.; Slaughter, G.W.; Koike, S.T. Phytophthora ramorum and Sudden Oak Death in California: I. Host Relationships. In General Technical Report PSW-GTR-184, Proceedings of the 5th Symposium on California Oak Woodlands, San Diego, CA, USA, 22–25 October 2001; Standiford, R., McCreary, D., Eds.; United States Department of Agriculture, Forest Service, Pacific Southwest Research Station: Albany, CA, USA, 2002; pp. 733–740. [Google Scholar]
- EPPO. Phytophthora ramorum. EPPO Bull. 2006, 36, 145–155. [Google Scholar]
- Hansen, E.; Sutton, W.; Parke, J.; Linderman, R. Phytophthora ramorum and Oregon Forest Trees—One Pathogen, Three Diseases. In Proceedings of the Sudden Oak Death Science Symposium, Monterey, CA, USA, 15–18 December 2002; p. 78. [Google Scholar]
- Davidson, J.M.; Werres, S.; Garbelotto, M.; Hansen, E.M.; Rizzo, D.M. Sudden oak death and associated diseases caused by Phytophthora ramorum. Plant Health Prog. 2003. [Google Scholar] [CrossRef]
- Hansen, E.M.; Kanaskie, A.; Prospero, S.; Mcwilliams, M.; Goheen, E.M.; Osterbauer, N.; Reeser, P.; Sutton, W. Epidemiology of Phytophthora ramorum in Oregon tanoak forests. Can. J. For. Res. 2008, 38, 1133–1143. [Google Scholar] [CrossRef]
- DiLeo, M.V.; Bostock, R.M.; Rizzo, D.M. Phytophthora ramorum does not cause physiologically significant systemic injury to California bay laurel, its primary reservoir host. Phytopathology 2009, 99, 1307–1311. [Google Scholar] [CrossRef] [PubMed]
- Davidson, J.M.; Wickland, A.C.; Patterson, H.A.; Falk, K.R.; Rizzo, D.M. Transmission of Phytophthora ramorum in mixed-evergreen forest in California. Phytopathology 2005, 95, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Werres, S.; Marwitz, R.; Man in’t Veld, W.A.; de Cock, A.W.; Bonants, P.J.; de Weerdt, M.; Themann, K.; Ilieva, E.; Baayen, R.P. Phytophthora ramorum sp. nov., a new pathogen on Rhododendron and Viburnum. Mycol. Res. 2001, 105, 1155–1165. [Google Scholar] [CrossRef]
- Davis, F.W.; Stoms, D.M.; Hollander, A.D.; Thomas, K.A.; Stine, P.A.; Odion, D.; Borchert, M.I.; Thorne, J.H.; Gray, M.V.; Walker, R.E. The California GAP Analysis Project—Final Report; University of California: Santa Barbara, CA, USA, 1998. [Google Scholar]
- Davidson, J.M.; Patterson, H.A.; Rizzo, D.M. Sources of inoculum for Phytophthora ramorum in a redwood forest. Phytopathology 2008, 98, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Meentemeyer, R.K.; Anacker, B.L.; Mark, W.; Rizzo, D.M. Early detection of emerging forest disease using dispersal estimation and ecological niche modeling. Ecol. Appl. 2008, 18, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Cobb, R.C.; Meentemeyer, R.K.; Rizzo, D.M. Apparent competition in canopy trees determined by pathogen transmission rather than susceptibility. Ecology 2010, 91, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Eyre, C.A.; Kozanitas, M.; Garbelotto, M. Population dynamics of aerial and terrestrial populations of Phytophthora ramorum in a California forest under different climatic conditions. Phytopathology 2013, 103, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Maywald, G.F.; Sutherst, R.W. A User's Guide to CLIMEX: A Computer Program for Comparing Climates in Ecology; Division of Entomology, CSIRO Australia: Canberra, Australia, 1985. [Google Scholar]
- Garbelotto, M.; Maddison, E.R.; Schmidt, D. SODmap and SODmap Mobile: Two tools to monitor the spread of sudden oak death. For. Phytophthoras 2014, 4. [Google Scholar] [CrossRef]
- Conrad, C.C.; Hilchey, K.G. A review of citizen science and community-based environmental monitoring: Issues and opportunities. Environ. Monit. Assess. 2011, 176, 273–291. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, J.L.; Shirk, J.; Bonter, D.; Bonney, R.; Crain, R.L.; Martin, J.; Phillips, T.; Purcell, K. The current state of citizen science as a tool for ecological research and public engagement. Front. Ecol. Environ. 2012, 10, 291–297. [Google Scholar] [CrossRef]
- Newman, G.; Wiggins, A.; Crall, A.; Graham, E.; Newman, S.; Crowston, K. The future of citizen science: Emerging technologies and shifting paradigms. Front. Ecol. Environ. 2012, 10, 298–304. [Google Scholar] [CrossRef]
- Zandbergen, P.A. Accuracy of iPhone locations: A comparison of assisted GPS, WiFi and cellular positioning. Trans. GIS 2009, 13, 5–25. [Google Scholar] [CrossRef]
- Hayden, K.; Ivors, K.; Wilkinson, C.; Garbelotto, M. TaqMan chemistry for Phytophthora ramorum detection and quantification, with a comparison of diagnostic methods. Phytopathology 2006, 96, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Mertikas, S.P. Error Distributions and Accuracy Measures in Navigation: An Overview; University of New Brunswick, Surveying Engineering Department: Fredericton, NB, Canada, 1985. [Google Scholar]
- Cormack, R.M. A review of classification. J. R. Stat. Soc. Ser. A. 1971, 134, 321–367. [Google Scholar] [CrossRef]
- Jones, H.G.; Vaughan, R.A. Remote Sensing of Vegetation: Principles, Techniques, and Applications; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Krishnamoorthy, K. Handbook of Statistical Distributions with Applications; CRC Press—Taylor & Francis Group: Boca Raton, FL, USA, 2016. [Google Scholar]
- Crawley, M.J. The R Book, 2nd ed.; John Wiley & Sons: Chichester, UK, 2013. [Google Scholar]
- Wagenmakers, E.J.; Farrell, S. AIC model selection using Akaike weights. Psychon. Bull. Rev. 2004, 11, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Grueber, C.E.; Nakagawa, S.; Laws, R.J.; Jamieson, I.G. Multimodel inference in ecology and evolution: Challenges and solutions. J. Evolut. Biol. 2011, 24, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Carsey, T.M.; Harden, J.J. Monte Carlo Simulation and Resampling: Methods for Social Science; Sage: Los Angeles, CA, USA, 2014. [Google Scholar]
- Cohen, J.A. Coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Hosmer, D.W.; Lemeshow, S. Applied Logistic Regression; Wiley: New York, NY, USA, 1989. [Google Scholar]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Mitchell, A. The Esri Guide to GIS Analysis: Geographic Patterns & Relationships; ESRI Press: Redlands, CA, USA, 1999; Volume 1. [Google Scholar]
- Silverman, B.W. Density Estimation for Statistics and Data Analysis; Chapman and Hall: London, UK, 1986. [Google Scholar]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S; Springer: New York, NY, USA, 2011. [Google Scholar]
- Conover, W.J.; Iman, R.L. Rank transformations as a bridge between parametric and nonparametric statistics. Am. Stat. 1981, 35, 124–129. [Google Scholar] [CrossRef]
- Blaker, H. Confidence curves and improved exact confidence intervals for discrete distributions. Can. J. Stat. 2000, 28, 783–798. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- PRISM Climate Group, Oregon State University. Available online: http://prism.oregonstate.edu (accessed on 15 January 2017).
- Jarvis, A.; Reuter, H.I.; Nelson, A.; Guevara, E. Hole-Filled SRTM for the Globe Version 4, Available from the CGIAR-CSI SRTM 90 m Database. 2008. Available online: http://srtm.csi.cgiar.org (accessed on 15 January 2017).
- Lamsal, S.; Cobb, R.C.; Cushman, J.H.; Meng, Q.; Rizzo, D.M.; Meentemeyer, R.K. Spatial estimation of the density and carbon content of host populations for Phytophthora ramorum in California and Oregon. For. Ecol. Manag. 2011, 262, 989–998. [Google Scholar] [CrossRef]
- Riley, S.J.; DeGloria, S.; Elliot, R.A. A Terrain ruggedness index that quantifies topographic heterogeneity. Intermt. J. Sci. 1999, 5, 23–27. [Google Scholar]
- De Reu, J.D.; Bourgeois, J.; Bats, M.; Zwertvaegher, A.; Gelorini, V.; Smedt, P.D.; Chu, W.; Antrop, M.; Maeyer, P.D.; Finke, P.; et al. Application of the topographic position index to heterogeneous landscapes. Geomorphology 2013, 186, 39–49. [Google Scholar] [CrossRef]
- Kennedy, H. Introduction to 3D Data: Modeling with ArcGIS 3D Analyst and Google Earth; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Wilcox, R.R.; Keselman, H.J.; Kowalchuk, R.K. Can tests for treatment group equality be improved? The bootstrap and trimmed means conjecture. Br. J. Math. Stat. Psychol. 1998, 51, 123–134. [Google Scholar] [CrossRef]
- Buckland, S.T. Monte Carlo confidence intervals. Biometrics 1984, 40, 811–817. [Google Scholar] [CrossRef]
- Pewsey, A.; Neuhäuser, M.; Ruxton, G.D. Circular Statistics in R; Oxford University Press: Oxford, UK, 2013. [Google Scholar]
- Rao, J.S. Large sample tests for the homogeneity of angular data. Sankhya Ser. B 1967, 28, 172–174. [Google Scholar]
- Mardia, K.V. A multi-sample uniform scores test on a circle and its parametric competitor. J. R. Stat. Soc. B Methodol. 1972, 102–113. [Google Scholar]
- Martin, F.N.; Tooley, P.W.; Blomquist, C. Molecular detection of Phytophthora ramorum, the causal agent of Sudden Oak Death in California, and two additional species commonly recovered from diseased plant material. Phytopathology 2004, 94, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Sweets, J.A. Measuring accuracy of diagnostic systems. Science 1988, 240, 1285–1293. [Google Scholar] [CrossRef]
- Garbelotto, M.; Davidson, J.M.; Ivors, K.; Maloney, P.E.; Hüberli, D.; Koike, S.T.; Rizzo, D.M. Non-oak native plants are main hosts for sudden oak death pathogen in California. Calif. Agric. 2003, 57, 18–23. [Google Scholar] [CrossRef]
- DiLeo, M.V.; Bostock, R.M.; Rizzo, D.M. Microclimate impacts survival and prevalence of Phytophthora ramorum in Umbellularia californica, a key reservoir host of sudden oak death in northern California forests. PLoS ONE 2014, 9, e98195. [Google Scholar] [CrossRef] [PubMed]
- Garbelotto, M.; Hayden, K.J. Sudden Oak Death: Interactions of the exotic oomycete Phytophthora ramorum with naïve North American hosts. Eukaryot. Cell 2012, 11, 1313–1323. [Google Scholar] [CrossRef] [PubMed]
- López-Escudero, F.J.; Blanco-López, M.A. Recovery of young olive trees from Verticillium dahliae. Eur. J. Plant Pathol. 2005, 113, 367–375. [Google Scholar] [CrossRef]
- Gordon, T.R.; Kirkpatrick, S.C.; Aegerter, B.J.; Fisher, A.J.; Storer, A.J.; Wood, D.L. Evidence for the occurrence of induced resistance to pitch canker, caused by Gibberella circinata (anamorph Fusarium circinatum), in populations of Pinus radiata. For. Pathol. 2010, 41, 227–232. [Google Scholar] [CrossRef]
- Midi, H.; Sarkar, S.K.; Rana, S. Collinearity diagnostics of binary logistic regression model. J. Interdiscip. Math. 2010, 13, 253–267. [Google Scholar] [CrossRef]
- Gibbon, J.D.; Pratt, J.W. P-values: Interpretation and methodogy. Am. Stat. 1975, 29, 20–25. [Google Scholar] [CrossRef]
- Kendrick, B. The Fifth Kingdom, 3rd ed.; Focus Publishing: Newburyport, MA, USA, 2000. [Google Scholar]
- Goralka, R.J.; Langenheim, J.H. Analysis of foliar monoterpenoid content in the California Bay Tree, Umbellularia californica, among populations across the distribution of the species. Biochem. Syst. Ecol. 1995, 23, 439–448. [Google Scholar] [CrossRef]
- Griffin, D.; Anchukaitis, K.J. How unusual is the 2012–2014 California drought? Geophys. Res. Lett. 2014, 41, 9017–9023. [Google Scholar] [CrossRef]
- Garrett, K.A.; Dendy, S.P.; Frank, E.E.; Rouse, M.N.; Travers, S.E. Climate change effects on plant disease: Genomes to ecosystems. Annu. Rev. Phytopathol. 2006, 44, 489–509. [Google Scholar] [CrossRef] [PubMed]
- Ansari, A.Q.; Loomis, W.E. Leaf temperatures. Am. J. Bot. 1959, 46, 713–717. [Google Scholar] [CrossRef]
- Fuchs, M. Infrared measurement of canopy temperature and detection of plant water-stress. Theor. Appl. Climatol. 1990, 42, 253–261. [Google Scholar] [CrossRef]
- Englander, L.; Browning, M.; Tooley, P.W. Growth and sporulation of Phytophthora ramorum in vitro in response to temperature and light. Mycologia 2006, 98, 365–373. [Google Scholar]
- Tooley, P.W.; Browning, M.; Berner, D. Recovery of Phytophthora ramorum following exposure to temperature extremes. Plant Dis. 2008, 92, 431–437. [Google Scholar] [CrossRef]
- Browning, M.; Englander, L.; Tooley, P.W.; Berner, D. Survival of Phytophthora ramorum hyphae after exposure to temperature extremes and various humidities. Mycologia 2008, 100, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Meentemeyer, R.K.; Moody, A.; Franklin, J. Landscape-scale patterns of shrub-species abundance in California chaparral—The role of topographically mediated resource gradients. Plant Ecol. 2001, 156, 19–41. [Google Scholar] [CrossRef]
- Diffenbaugh, N.S.; Pal, J.S.; Giorgi, F.; Gao, X. Heat stress intensification in the Mediterranean climate change hotspot. Geophys. Res. Lett. 2007, 34, L11706. [Google Scholar] [CrossRef]
- Di Castri, F. An ecological overview of the five regions of the world with a Mediterranean climate. In Biogeography of Mediterranean Invasions; Groves, R.H., di Castri, F., Eds.; Cambridge University Press: Cambridge, UK, 1991; pp. 3–15. [Google Scholar]
- Mark, D.M.; Aronson, P.B. Scale-dependent fractal dimensions of topographic surfaces: An empirical investigation, with applications in geomorphology and computer mapping. Math. Geol. 1984, 16, 671–683. [Google Scholar] [CrossRef]
- Swanson, F.J.; Kratz, T.K.; Caine, N.; Woodmansee, R.G. Landform effects on ecosystem patterns and processes. BioScience 1988, 38, 92–98. [Google Scholar] [CrossRef]
- Catani, F.; Segoni, S.; Falorni, G. An empirical geomorphology-based approach to the spatial prediction of soil thickness at catchment scale. Water Resour. Res. 2010, 46, W05508. [Google Scholar] [CrossRef]
- Swiecki, T.J.; Bernhardt, E.A. Increasing distance from California bay laurel reduces the risk and severity of Phytophthora ramorum canker in coast live oak. In General Technical Report PSW-GTR-214, Proceedings of the Sudden Oak Death Third Science Symposium, Santa Rosa, California, 5-9 March 2007; Frankel, S.J., Kliejunas, J.T., Palmieri, K.M., Eds.; United States Department of Agriculture, Forest Service, Pacific Southwest Research Station: Albany, CA, USA, 2008; pp. 181–194. [Google Scholar]
- Hüberli, D.; Garbelotto, M. Phytophthora ramorum is a generalist plant pathogen with differences in virulence between isolates from infectious and dead-end hosts. For. Pathol. 2012, 42, 8–13. [Google Scholar] [CrossRef]
- Giordano, L.; Gonthier, P.; Lione, G.; Capretti, P.; Garbelotto, M. The saprobic and fruiting abilities of the exotic forest pathogen Heterobasidion irregulare may explain its invasiveness. Biol. Invasions 2014, 16, 803–814. [Google Scholar] [CrossRef]
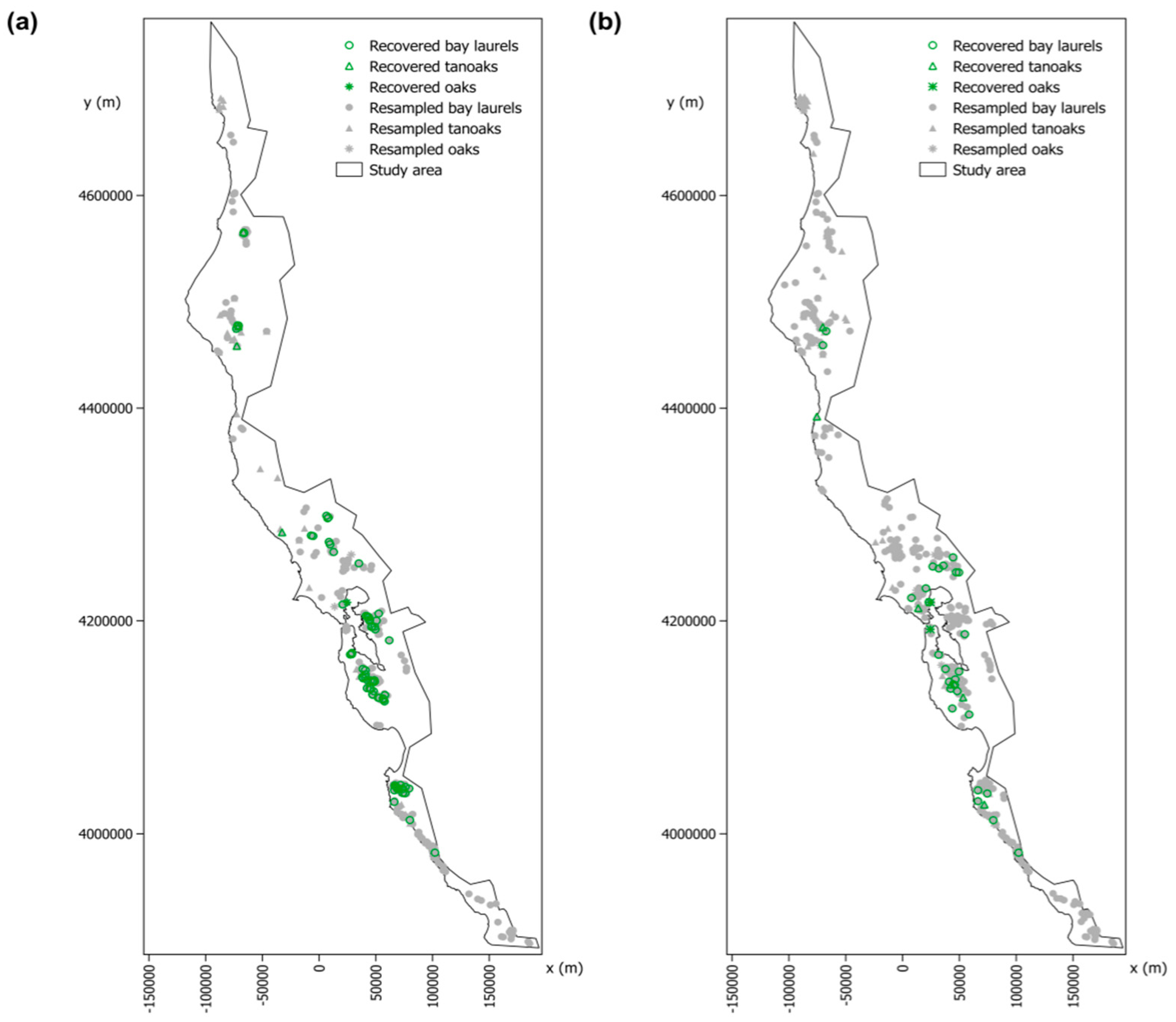

| Species | Scenario | Trees Infected at Least Once by P. ramorum | Recovered Trees | Recovered Trees (%) | 95% CI Lower Bound (%) | 95% CI Upper Bound (%) |
|---|---|---|---|---|---|---|
| Bay laurel | 10 m | 359 | 116 | 32.3 | 27.5 | 37.3 |
| 500 m | 82 | 27 | 32.9 | 23.4 | 43.8 | |
| Tanoak | 10 m | 43 | 4 | 9.3 | 3.2 | 21.5 |
| 500 m | 36 | 7 | 19.4 | 8.8 | 35.8 | |
| Oaks | 10 m | 6 | 1 | 16.7 | 0.9 | 59.4 |
| 500 m | 8 | 2 | 25.0 | 4.6 | 64.1 | |
| Overall species | 10 m | 408 | 121 | 29.7 | 25.3 | 34.3 |
| 500 m | 126 | 36 | 28.6 | 21.2 | 37.2 |
| Scenario | W | W p-Value | R1 | R1 p-Value | R2 | R2 p-Value |
|---|---|---|---|---|---|---|
| 10 m | 1.974 | 0.373 | 1.026 | 0.311 | 0.178 | 0.673 |
| 500 m | 0.236 | 0.888 | 0.428 | 0.513 | 0.006 | 0.938 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lione, G.; Gonthier, P.; Garbelotto, M. Environmental Factors Driving the Recovery of Bay Laurels from Phytophthora ramorum Infections: An Application of Numerical Ecology to Citizen Science. Forests 2017, 8, 293. https://doi.org/10.3390/f8080293
Lione G, Gonthier P, Garbelotto M. Environmental Factors Driving the Recovery of Bay Laurels from Phytophthora ramorum Infections: An Application of Numerical Ecology to Citizen Science. Forests. 2017; 8(8):293. https://doi.org/10.3390/f8080293
Chicago/Turabian StyleLione, Guglielmo, Paolo Gonthier, and Matteo Garbelotto. 2017. "Environmental Factors Driving the Recovery of Bay Laurels from Phytophthora ramorum Infections: An Application of Numerical Ecology to Citizen Science" Forests 8, no. 8: 293. https://doi.org/10.3390/f8080293





