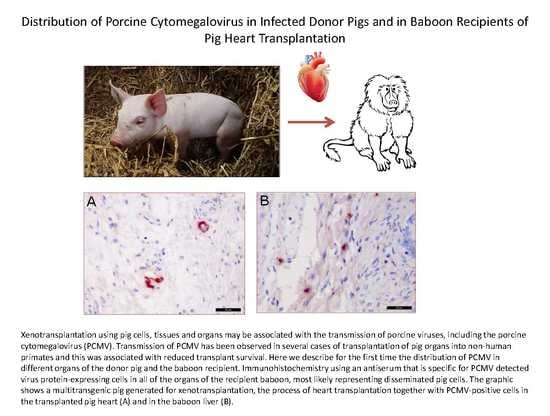
Distribution of Porcine Cytomegalovirus in Infected Donor Pigs and in Baboon Recipients of Pig Heart Transplantation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Transplantations
2.2. Ethics Statement
2.3. Testing for PCMV, HEV and BaCMV, PCR Methods
2.4. Determination of the PCMV Copy Number
2.5. Determination of the BaCMV Copy Number
2.6. PBMC Incubation
2.7. PCMV and HEV Antigens Used for Western Blot Analyses
2.8. Testing for PCMV, HEV and PLHV
2.9. Immunohistochemistry (IHC)
2.10. Sequence Alignment
3. Results
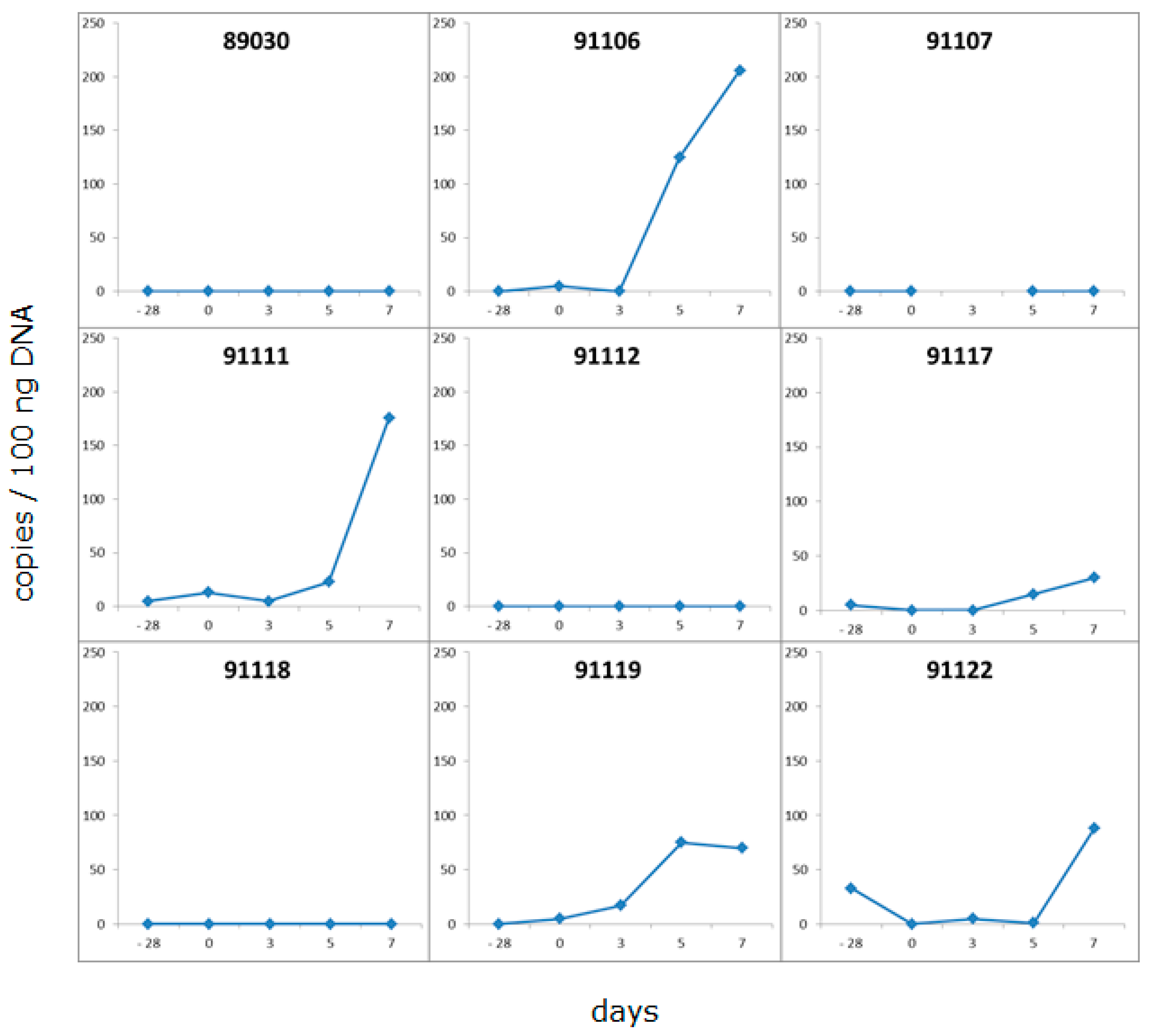
3.1. Time Schedule of the Transplantation Experiments and Testing for PCMV
3.2. Detection of PCMV in Donor Pig and Baboon Recipients
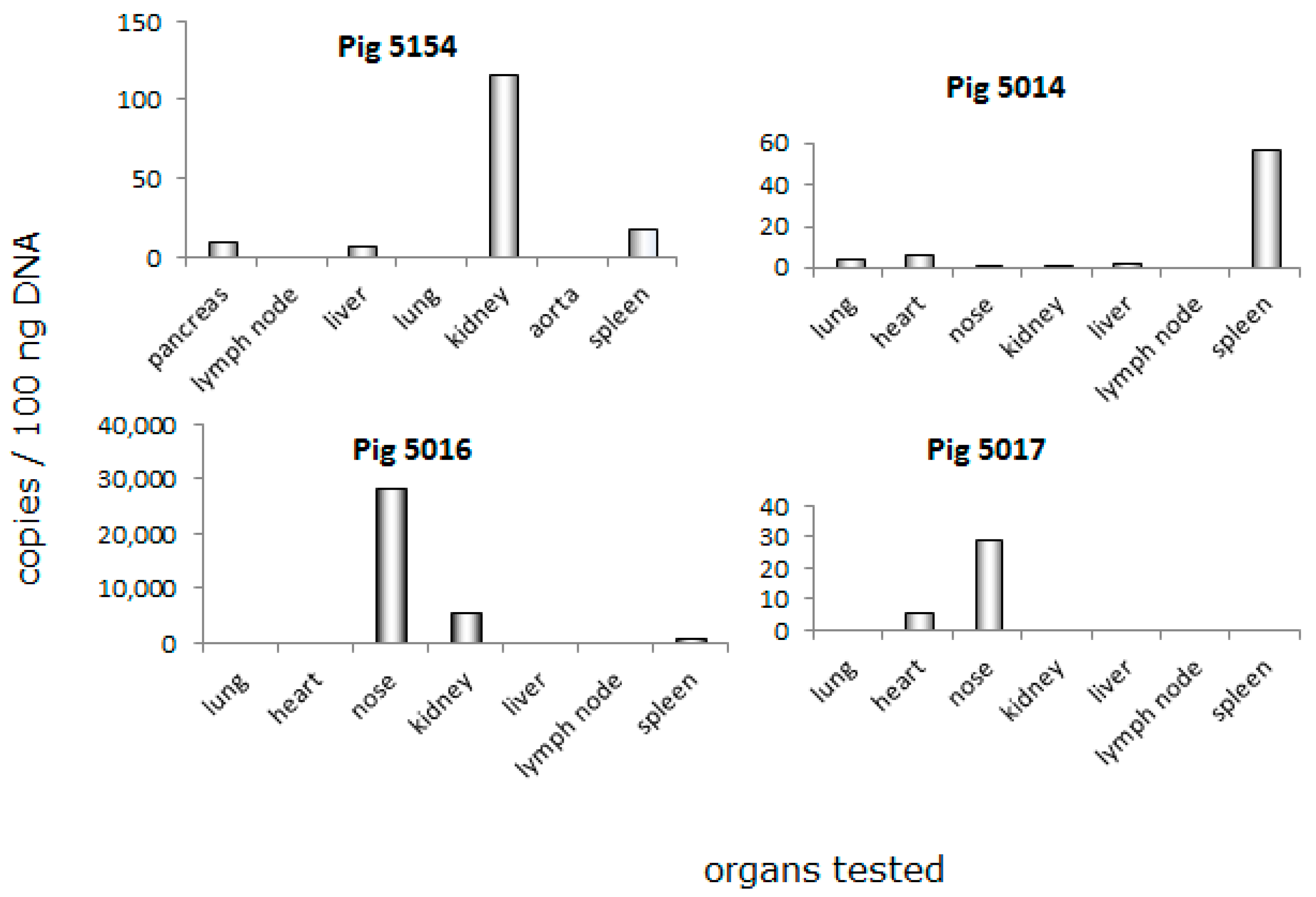
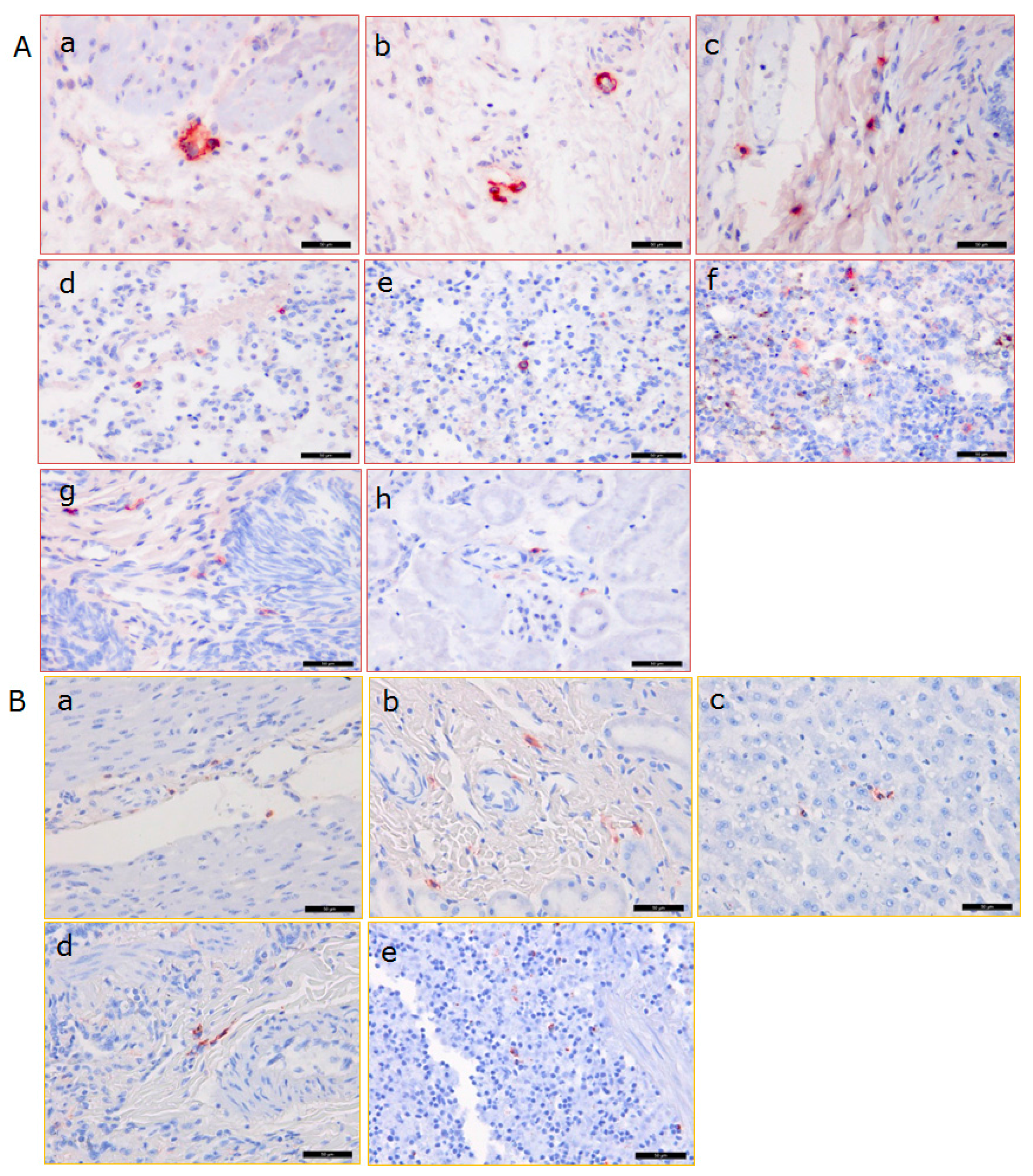
3.3. Distribution of PCMV in Pig Organs
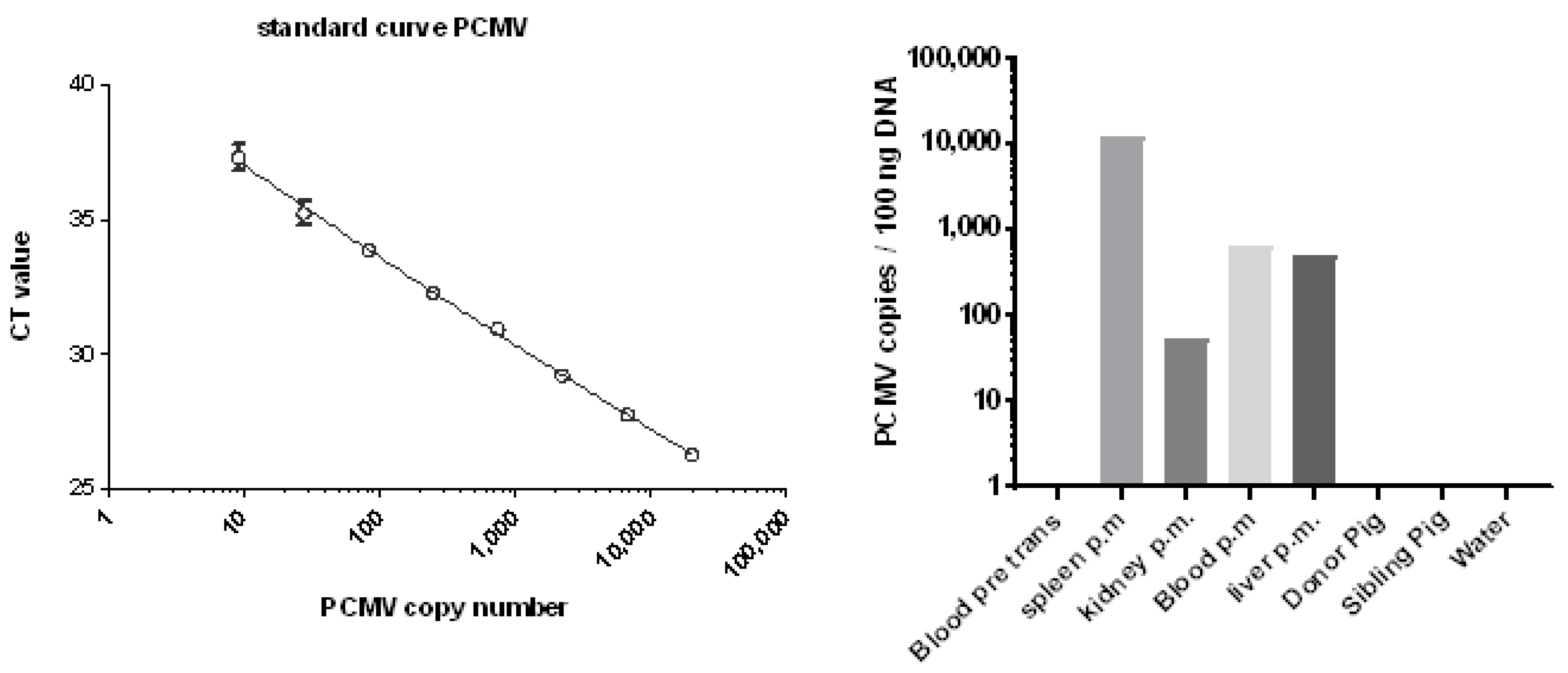
3.4. Development of a New Detection Strategy Using Incubated PBMC
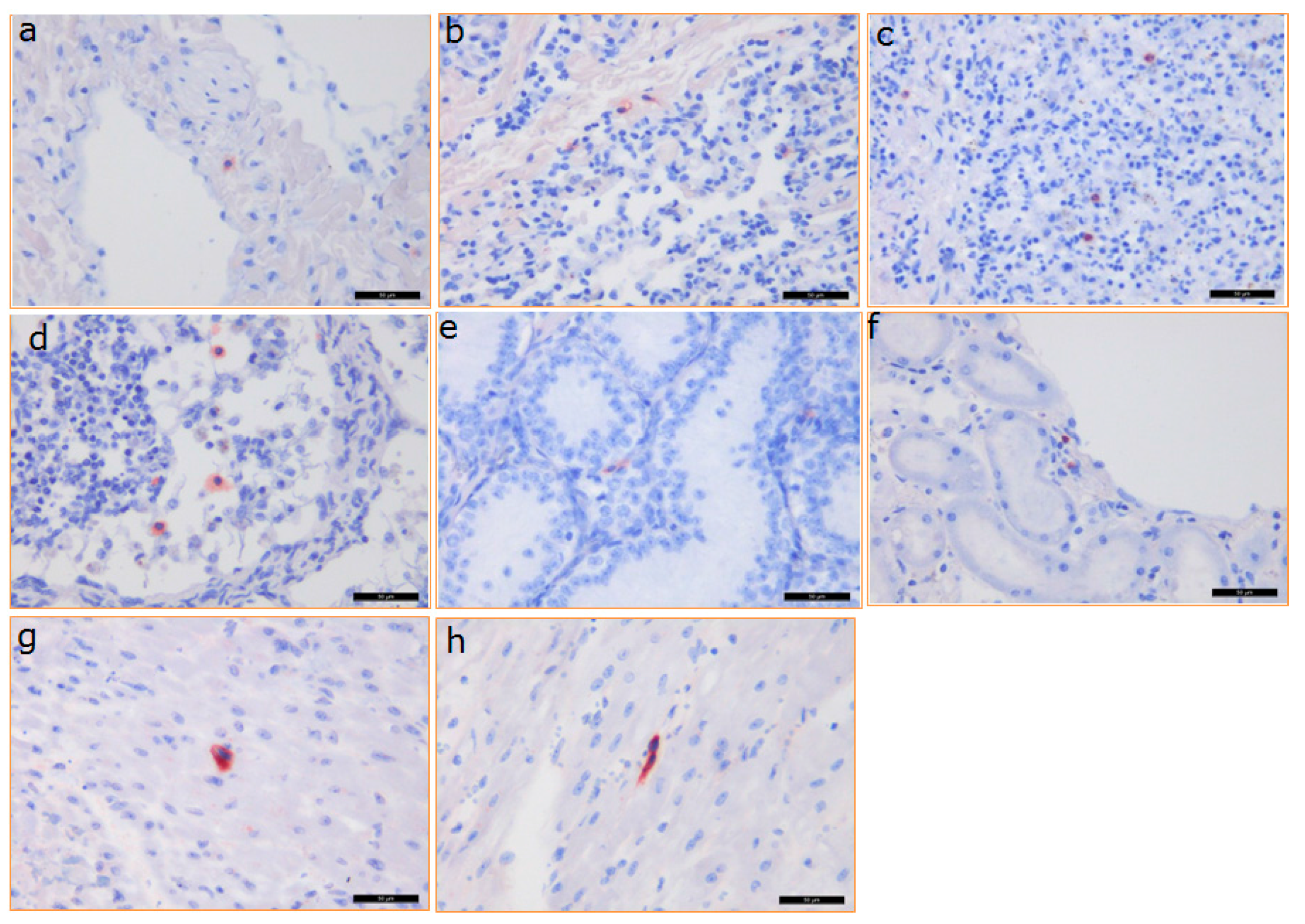
3.5. Distribution of PCMV in Baboon Organs
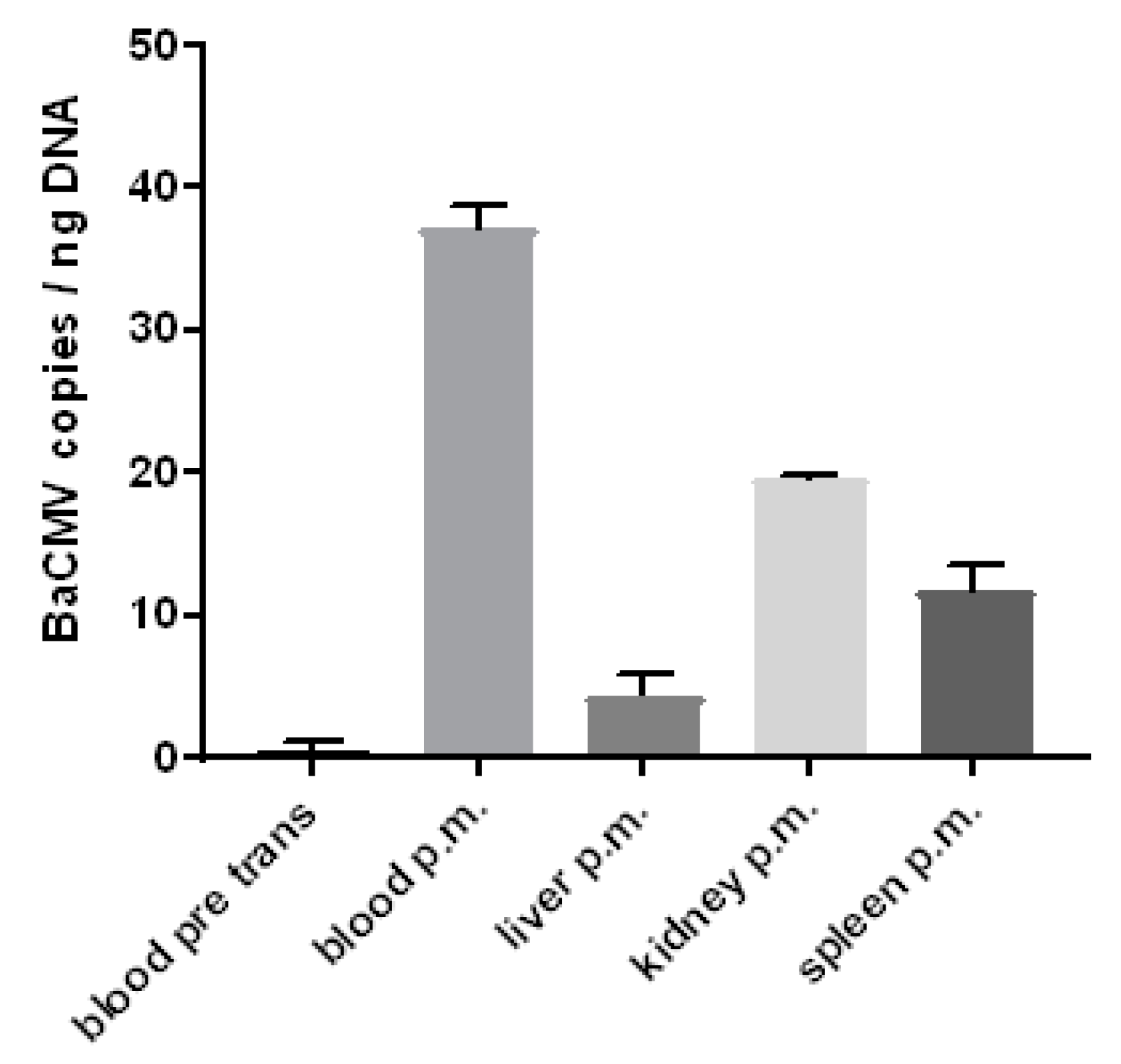
3.6. Detection of the Baboon Cytomegalovirus (BaCMV) in the Recipient Baboons
3.7. Immunological Cross-Reactivity between PCMV and BaCMV
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ekser, B.; Ezzelarab, M.; Hara, H.; van der Windt, D.J.; Wijkstrom, M.; Bottino, R.; Trucco, M.; Cooper, D.K. Clinical xenotransplantation, the next medical revolution? Lancet 2012, 379, 672–683. [Google Scholar] [CrossRef]
- Tacke, F.; Kroy, D.C.; Barreiros, A.P.; Neumann, U.P. Liver transplantation in Germany. Liver Transplant. 2016, 22, 1136–1142. [Google Scholar] [CrossRef] [PubMed]
- Niemann, H.; Petersen, B. The production of multi-transgenic pigs, update and perspectives for xenotransplantation. Transgen. Res. 2016, 25, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Klymiuk, N.; Aigner, B.; Brem, G.; Wolf, E. Genetic modification of pigs as organ donors for xenotransplantation. Mol. Reprod. Dev. 2010, 77, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Fishman, J.A.; Patience, C. Xenotransplantation, infectious risk revisited. Am. J. Transplant. 2004, 4, 1383–1390. [Google Scholar] [CrossRef] [PubMed]
- Fishman, J.A.; Scobie, L.; Takeuchi, Y. Xenotransplantation-associated infectious risk, a WHO consultation. Xenotransplantation 2012, 19, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Edington, N. Porcine cytomegalovirus. Dis. Swine 1986, 138, 330–336. [Google Scholar]
- Edington, N.; Broad, S.; Wrathall, A.E.; Done, J.T. Superinfection with porcine cytomegalovirus initiate infection. Vet. Microbiol. 1988, 16, 189–193. [Google Scholar] [CrossRef]
- Mueller, N.J.; Kuwaki, K.; Knosalla, C.; Dor, F.J.; Gollackner, B.; Wilkinson, R.A.; Arn, S.; Sachs, D.H.; Cooper, D.K.; Fishman, J.A. Early weaning of piglets fails to exclude porcine lymphotropic herpesvirus. Xenotransplantation 2005, 12, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Tasaki, M.; Sekijima, M.; Wilkinson, R.A.; Villani, V.; Moran, S.G.; Cormack, T.A.; Hanekamp, I.M.; Hawley, R.J.; Arn, J.S.; et al. Porcine cytomegalovirus infection is associated with early rejection of kidney grafts in a pig to baboon xenotransplantation model. Transplantation 2014, 98, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Sekijima, M.; Waki, S.; Sahara, H.; Tasaki, M.; Wilkinson, R.A.; Villani, V.; Shimatsu, Y.; Nakano, K.; Matsunari, H.; Nagashima, H. Results of life-supporting galactosyltransferase knockout kidneys in cynomolgus monkeys using two different sources of galactosyltransferase knockout swine. Transplantation 2014, 98, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.A.; Fryer, J.F.L.; Tucker, A.W.; McArdle, P.D.; Hughes, A.E.; Emery, V.C.; Griffiths, P.D. Porcine cytomegalovirus in pigs being bred for xenograft organs, progress towards control. Xenotransplantation 2003, 10, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Tucker, A.W.; Galbraith, D.; McEwan, P.; Onions, D. Evaluation of porcine cytomegalovirus as a potential zoonotic agent in xenotransplantation. Transplant. Proc. 1999, 31, 915. [Google Scholar] [CrossRef]
- Denner, J.; Mueller, N.J. Preventing transfer of infectious agents. Int. J. Surg. 2015, 23, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.J.; Kuwaki, K.; Dor, F.J.; Knosalla, C.; Gollackner, B.; Wilkinson, R.A.; Sachs, D.H.; Cooper, D.K.; Fishman, J.A. Reduction of consumptive coagulopathy using porcine cytomegalovirus-free cardiac porcine grafts in pig-to-primate xenotransplantation. Transplantation 2004, 78, 1449–1453. [Google Scholar] [CrossRef] [PubMed]
- Goltz, M.; Widen, F.; Banks, M.; Belak, S.; Ehlers, B. Characterization of the DNA polymerase loci of porcine cytomegalovirus from diverse geographical origins. Virus Genes 2000, 21, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Fryer, J.F.L.; Griffiths, P.D.; Fishman, J.A.; Emery, V.C.; Clark, D.A. Quantitation of porcine cytomegalovirus in pig tissues by PCR. J. Clin. Microbiol. 2001, 39, 1155–1156. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.A.; Morozov, A.V.; Denner, J. New nested and real-time PCR systems for porcine cytomegalovirus (PCMV) detection and quantification. Arch. Virol. 2016, 161, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Widen, B.F.; Lowings, J.P.; Belak, S.; Banks, M. Development of a PCR system for porcine cytomegalovirus detection and determination of the putative partial sequence of its DNA polymerase gene. Epidemiol. Infect. 1999, 123, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Hamel, A.L.; Lin, L.; Sachvie, C.; Grudeski, E.; Nayar, G.P. PCR assay for detecting porcine cytomegalovirus. J. Clin. Microbiol. 1999, 37, 3767–3768. [Google Scholar] [PubMed]
- Plotzki, E.; Keller, M.; Ivanusic, D.; Denner, J. A new Western blot assay for the detection of porcine cytomegalovirus (PCMV). J. Immunol. Methods 2016, 437, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhu, L.; Shi, X.; Xu, Z.; Mei, M.; Xu, W.; Zhou, Y.; Guo, W.; Wang, X. Indirect-blocking ELISA for detecting antibodies against glycoprotein B (gB) of porcine cytomegalovirus (PCMV). J. Virol. Methods. 2012, 186, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, U.; Holzer, A.; Ivanusic, D.; Plotzki, P.; Hengel, H.; Neipel, F.; Denner, J. Antibody cross-reactivity between porcine cytomegalovirus (PCMV) and human herpesvirus-6 (HHV-6). Viruses 2017, 9, 317. [Google Scholar] [CrossRef] [PubMed]
- Denner, J. Xenotransplantation and porcine cytomegalovirus. Xenotransplantation 2015, 22, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.A.; Abicht, J.M.; Reichart, B.; Mayr, T.; Guethoff, S.; Denner, J. Active replication of porcine cytomegalovirus (PCMV) following transplantation of a pig heart into a baboon despite undetected virus in the donor pig. Ann. Virol. Res. 2016, 2, 1018. [Google Scholar]
- Lower, R.R.; Shumway, N.E. Studies on the orthotopic homotransplantation of the canine heart. Surg. Forum. 1960, 11, 18–19. [Google Scholar] [PubMed]
- Morozov, V.A.; Ludwig, S.; Ludwig, B.; Rotem, A.; Barkai, U.; Bornstein, S.; Denner, J. Islet Cell Transplantation from Göttingen Minipigs to Cynomolgus Monkeys, Analysis of Virus Safety. Xenotransplantation 2016, 23, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.A.; Plotzki, E.; Rotem, A.; Barkai, U.; Denner, J. Extended Microbiological Characterization of Göttingen Minipigs, Porcine Cytomegalovirus and Other Viruses. Xenotransplantation 2016, 227, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.A.; Morozov, A.V.; Rotem, A.; Barkai, U.; Bornstein, S.; Denner, J. Extended Microbiological Characterization of Göttingen Minipigs in the Context of Xenotransplantation, Detection and Vertical Transmission of Hepatitis E Virus. PLoS ONE 2015, 10, e0139893. [Google Scholar] [CrossRef] [PubMed]
- Abicht, J.M.; Mayr, T.A.; Reichart, B.; Plotzki, E.; Güthoff, S.; Falkenau, A.; Kind, A.; Denner, J. Hepatic Failure After Pig Heart Transplantation Into a Baboon, No Involvement of Porcine Hepatitis E Virus. Ann. Transplant. 2016, 21, 12–16. [Google Scholar] [PubMed]
- Mueller, N.J.; Barth, R.N.; Yamamoto, S.; Kitamura, H.; Patience, C.; Yamada, K.; Cooper, D.K.; Sachs, D.H.; Kaur, A. Activation of cytomegalovirus in pig-to-primate organ xenotransplantation. J. Virol. 2002, 76, 4734–4764. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, B.; Robert Koch Institute, Berlin, Germany. Unpublished work. 2002.
- Chmielewicz, B.; Goltz, M.; Franz, T.; Bauer, C.; Brema, S.; Ellerbrok, H.; Beckmann, S.; Rziha, H.J.; Lahrmann, K.H.; Romero, C.; et al. A novel porcine gammaherpesvirus. Virology 2003, 308, 317–329. [Google Scholar] [CrossRef]
- Duvigneau, J.C.; Hartl, R.T.; Groiss, S.; Gemeiner, M. Quantitative simultaneous multiplex real-time PCR for the detection of porcine cytokines. J. Immunol. Methods 2005, 30, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.J.; Hill, V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 2006, 131, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.J.; Livingston, C.; Knosalla, C.; Barth, R.N.; Yamamoto, S.; Gollackner, B.; Dor, F.J.; Buhler, L.; Sachs, D.H.; Yamada, K.; et al. Activation of porcine cytomegalovirus, but not porcine lymphotropic herpesvirus, in pig-to-baboon xenotransplantation. J. Infect. Dis. 2004, 189, 1628–1633. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Zeng, N.; Zhou, L.; Ge, X.; Guo, X.; Yang, H. Genomic organization and molecular characterization of porcine cytomegalovirus. Virology 2014, 460–461, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Grote, A.; Hiller, K.; Scheer, M.; Münch, R.; Nörtemann, B.; Hempel, D.C.; Jahn, D. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucl. Acids Res. 2005, 33, W526–W531. [Google Scholar] [CrossRef] [PubMed]
- Plotzki, E.; Keller, M.; Ehlers, B.; Denner, J. Immunological Methods for the Detection of Porcine Lymphotropic Herpesviruses (PLHV). J. Virol. Methods 2016, 233, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, M.; Shibahara, T.; Miyazaki, A.; Tajima, T.; Shimizu, S.; Kabali, E.; Takano, Y.; Sasaki, Y.; Kubo, M. In situ hybridization and immunohistochemistry for the detection of porcine cytomegalovirus. J. Virol. Methods 2012, 179, 272–275. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit, a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Fiebig, U.; Runge, C.; Denner, J.; Robert Koch Institute, Berlin, Germany. Unpublished work. 2017.
- Shibahara, T.; National Institute of Animal Health, Tsukuba, Japan. Unpublished work. 2017.
- Morozov, V.A.; Heinrichs, G.; Denner, J. Effective Detection of Porcine Cytomegalovirus Using Non-Invasively Taken Samples from Piglets. Viruses 2017, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Neumann, E.J.; Ramirez, A.; Schwartz, K.J. Swine Disease Manual, 4th ed.; AASV: Ames, IA, USA, 2010; pp. 53–54. [Google Scholar]
- Guedes, M.I.M.C.; Risdah, I.J.M.; Wiseman, B.; Molitor, T.W. Reactivation of porcine cytomegalovirus through allogenic stimulation. J. Clin. Microbiol. 2004, 42, 1756–1758. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.J. Herpesvirus systematics. Vet. Microbiol. 2010, 143, 52–69. [Google Scholar] [CrossRef] [PubMed]
- Jurak, I.; Brune, W. Induction of apoptosis limits cytomegalovirus cross-species infection. EMBO J. 2006, 25, 2634–2642. [Google Scholar] [CrossRef] [PubMed]
- Whitteker, J.L.; Dudani, A.K.; Tackaberry, E.S. Human fibroblasts are permissive for porcine cytomegalovirus in vitro. Transplantation 2008, 86, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Blewett, E.; White, G.; Saliki, J.; Eberle, R. Isolation and characterization of an endogenous cytomegalovirus (BaCMV) from baboons. Arch. Virol. 2001, 146, 1723–1738. [Google Scholar] [CrossRef] [PubMed]
- Ross, T.G.; Rogers, R.P.; Elfrink, N.; Bray, N.; Blewett, E.L. Detection of baboon cytomegalovirus (BaCMV) by PCR using primers directed against the glycoprotein B gene. J. Virol. Methods 2005, 125, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Michaels, M.G.; Jenkins, F.J.; St Geroge, K.; Nalesnik, M.A.; Starzl, T.E.; Rinaldo, C.R. Detection of Infectious Baboon Cytomegalovirus after Baboon-to-Human Liver Xenotransplantation. J. Virol. 2001, 75, 2825–2828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaels, M.G.; Alcendor, D.; St George, K.; Rinaldo, C.R.; Ehrlich, G.D.; Becich, M.J.; Hayward, G.S. Distinguishing baboon cytomegalovirus from human cytomegalovirus, Importance for xenotransplantation. J. Infect. Dis. 1997, 176, 1476–1483. [Google Scholar] [CrossRef] [PubMed]

| Primers Used for PCR | Sequence 5′-3′ | Nucleotide Position | Accession Number | Reference |
|---|---|---|---|---|
| PLHV-1,-2 (747) fw PLHV-1,-2 (747) rev | CAYGGTAGTATTTATTCAGACA GATATCCTGGTACATTGGAAAG | 21,146–21,167 21,488–21,467 | AY170317.1 | Ehlers B., 2002 [32] |
| PLHV-3 (905) fw PLHV-3 (905) rev | ACAAGAGCCTTAGGGTTCCAAACT GTGTCCAGTGTTGTAATGGATGCC | 13,472–13,495 13,727–13,704 | AY170316.1 | Chmielewicz et al., 2003 [33] |
| Primers and probes used for real-time PCR | ||||
| pcyclophilin real fw pcyclophilin real rev pcyclophilin probe | TGCTTTCACAGAATAATTCCAGGATTTA GACTTGCCACCAGTGCCATTA Cy5-TGCCAGGGTGGTGACTTCACACGCC-BHQ | 131–158 187–207 161–185 | AY008846 | Duvigneau et al., 2005 [34] |
| pGAPDH fw pGAPDH rev pGAPDH probe | ACATGGCCTCCAAGGAGTAAGA GATCGAGTTGGGGCTGTGACT HEX-CCACCAACCCCAGCAAGAG-BHQ1 | 1040–1062 1188–1168 1114–1132 | NM_001206359.1 | Duvigneau et al., 2005 [34] |
| JVHEVF JVHEVR JVHEVProbe | GGTGGTTTCTGGGGTGAC TGATTCTCAGCCCTTCGC FAM-TGATTCTCAGCCCTTCGC-BHQ | 5283–5300 5352–5335 5306–5323 | KT633715.1 | Jothikumar et al., 2006 [35] |
| PCMV real fw PCMV real rev PCMV probe | ACGAGAAAGATATTCTGACGGTGCA TCTAGACGAAAGGACATTGTTGATA 6FAM-CAGGGCGGCGGTCGAGCTC-TAMRA | 45,962–45,938 45,622–45,646 45,246–45,229 | KF017583.1 | Mueller et al., 2004 [36] |
| BaCMV real fw BaCMV real rev BaCMV probe | 5′ GTTTAGGGAACCGCCATTCTG 3′ 5′ GTATCCGCGTTCCAATGCA 3′ 5′ 6FAM-TCCAGCCTCCATAGCCGGGAAGG-BBQ 3′ | 64,375–64,396 64,464–64,483 64,437–64,460 | KR351281 | Mueller et al., 2002 [31] |
| BaCCR5 real fw BaCCR5 real rev BaCCR5 probe | 5′ TACCTGCTCAACCTGGCCAT 3′ 5′ TTCCAAAGTCCACTGGGC 3′ HEX-TTTCCTTCTTACTGTCCCCTTCTGGGCTC-BHQ 3′ | Mueller et al., 2002 [31] | ||
| Pig Number | Born | Gender | Material Collected (Date) | PCMV (Real-Time PCR) | HEV (Real-Time RT PCR) | PLHV1/2 (PCR) | PLHV3 (PCR) | PCMV (Western Blot) * | PLHV (Western Blot) ** | HEV (Western Blot) *** |
|---|---|---|---|---|---|---|---|---|---|---|
| 5154 | 1 July 2016 | male | 7 September 2016 | no ct | no ct | no ct | no ct | negative | negative | negative |
| 5160 | 1 July 2016 | female | 7 September 2016 | no ct | no ct | no ct | no ct | negative | negative | negative |
| 5160 | 3 February 2017 | no ct | no ct | no ct | no ct | negative | negative | negative | ||
| 5157 | 1 July 2016 | female | 26 October 2016 | no ct | no ct | no ct | no ct | negative | negative | negative |
| Organ | Baboons | ||
|---|---|---|---|
| 57 | 64 | Control 43 | |
| Liver | + | + | + * |
| Spleen | + | ++ | + * |
| Kidney | + | + | + * |
| Pig heart | ++ | ++ | + * |
| Lung | + | + | + * |
| Testis | +++ | + | n.t. |
| Lymph node | +++ | +++ | n.t. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiebig, U.; Abicht, J.-M.; Mayr, T.; Längin, M.; Bähr, A.; Guethoff, S.; Falkenau, A.; Wolf, E.; Reichart, B.; Shibahara, T.; et al. Distribution of Porcine Cytomegalovirus in Infected Donor Pigs and in Baboon Recipients of Pig Heart Transplantation. Viruses 2018, 10, 66. https://doi.org/10.3390/v10020066
Fiebig U, Abicht J-M, Mayr T, Längin M, Bähr A, Guethoff S, Falkenau A, Wolf E, Reichart B, Shibahara T, et al. Distribution of Porcine Cytomegalovirus in Infected Donor Pigs and in Baboon Recipients of Pig Heart Transplantation. Viruses. 2018; 10(2):66. https://doi.org/10.3390/v10020066
Chicago/Turabian StyleFiebig, Uwe, Jan-Michael Abicht, Tanja Mayr, Matthias Längin, Andrea Bähr, Sonja Guethoff, Almuth Falkenau, Eckhard Wolf, Bruno Reichart, Tomoyuki Shibahara, and et al. 2018. "Distribution of Porcine Cytomegalovirus in Infected Donor Pigs and in Baboon Recipients of Pig Heart Transplantation" Viruses 10, no. 2: 66. https://doi.org/10.3390/v10020066