High Whole-Genome Sequence Diversity of Human Papillomavirus Type 18 Isolates
Abstract
:1. Introduction
2. Materials and Methods
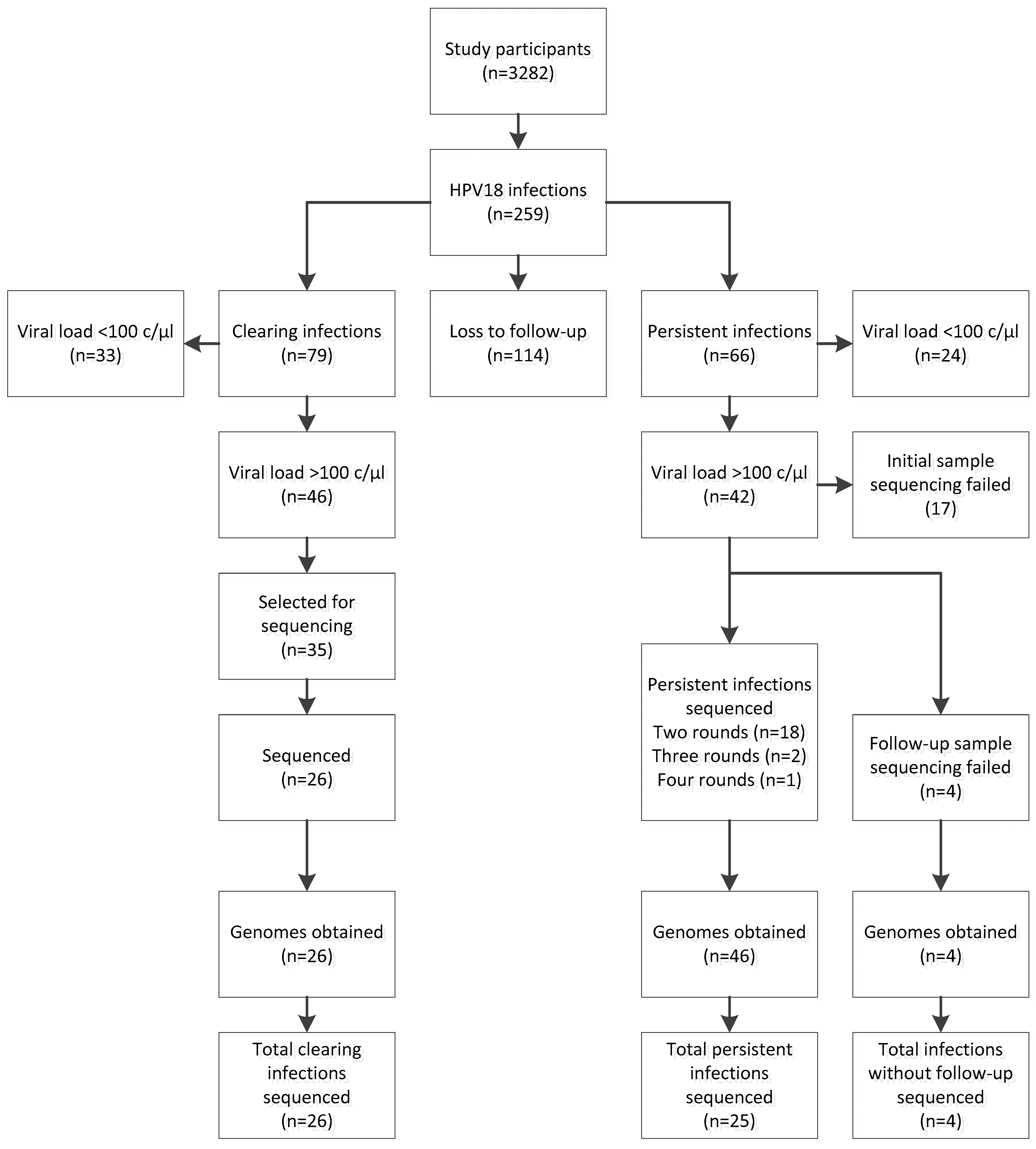
2.1. Study Design
2.2. DNA Purification and HPV Detection, Genotyping and Quantification
2.3. Sample Selection, Long-Template PCR and Sequencing
2.4. Phylogenetic and SNP Analysis
2.5. Statistical Analysis
2.6. Accession Numbers
3. Results
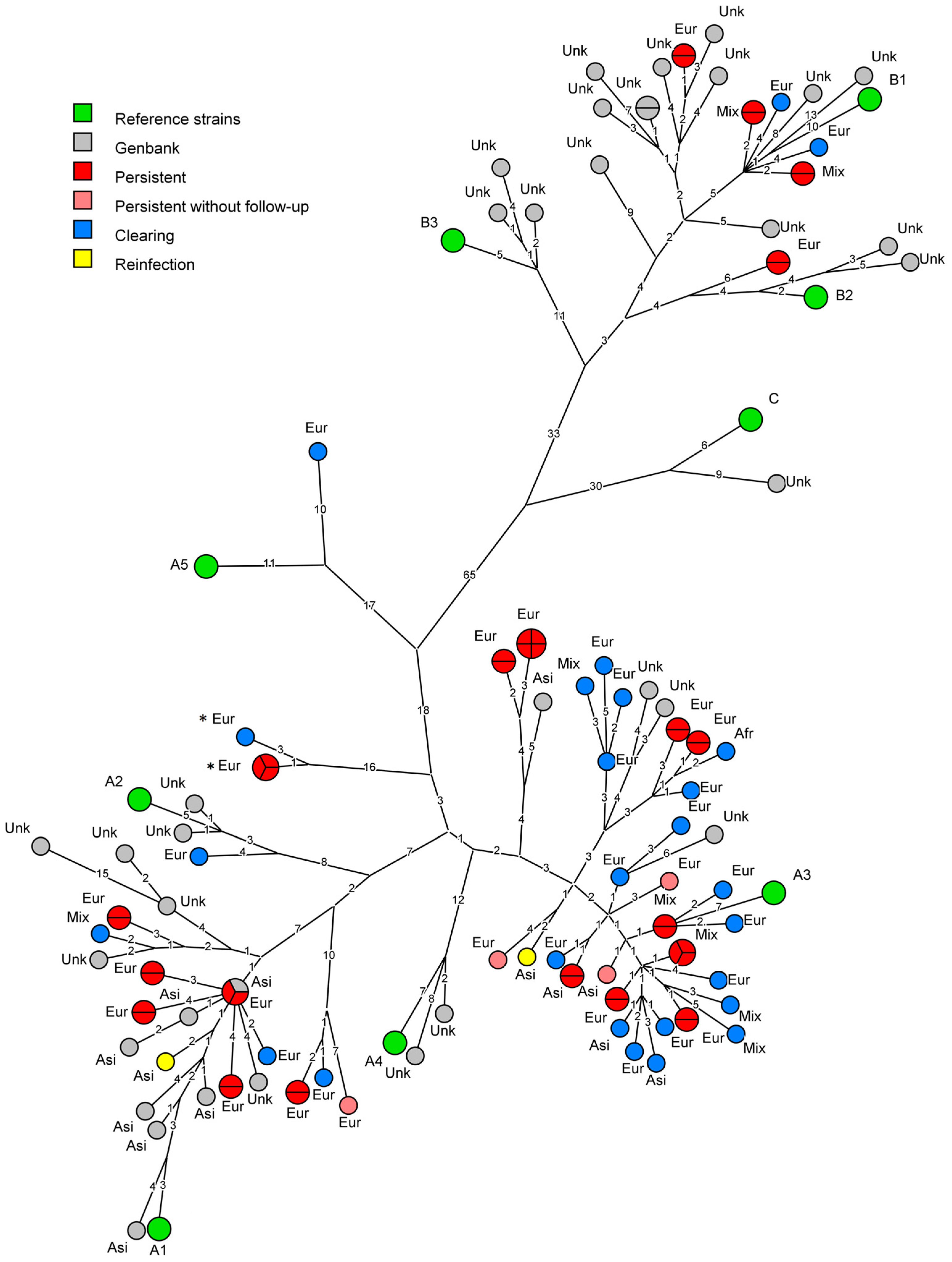
3.1. Phylogeny
3.2. Analysis of Persistent Infections
3.3. Comparison to Currently Available Data
3.4. Ethnicity
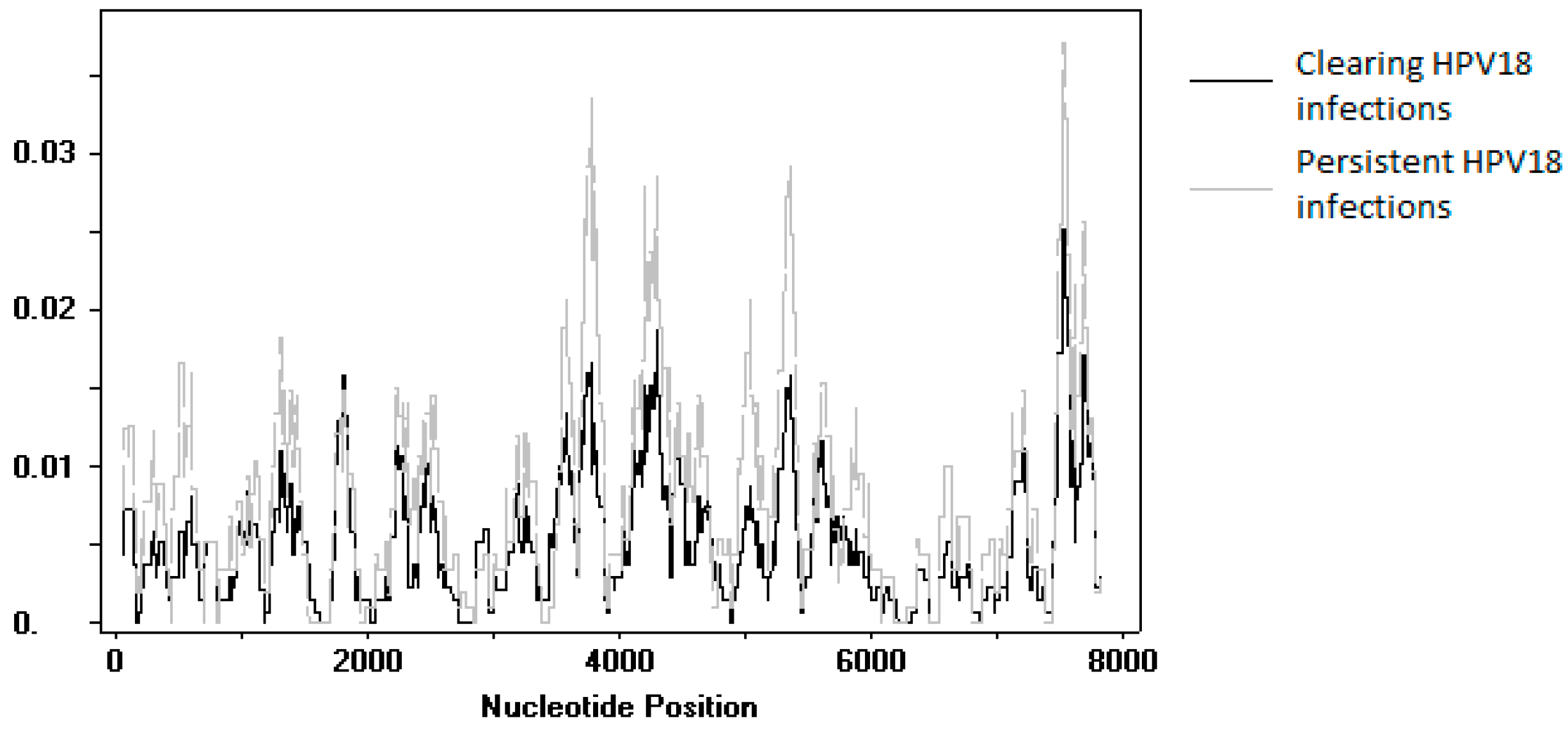
3.5. SNP Analysis
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.; Peto, J.; Meijer, C.J.; Munoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Biological agents. Volume 100 B. A review of human carcinogens. IARC Monogr. Eval. Carcinog. Risks Hum. 2012, 100, 1–441. [Google Scholar]
- De Sanjose, S.; Quint, W.G.; Alemany, L.; Geraets, D.T.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.R.; et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef]
- Burk, R.D.; Harari, A.; Chen, Z. Human papillomavirus genome variants. Virology 2013, 445, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.F.; Koutsky, L.A.; Hildesheim, A.; Galloway, D.A.; Wheeler, C.M.; Winer, R.L.; Ho, J.; Kiviat, N.B. Risk for high-grade cervical intraepithelial neoplasia associated with variants of human papillomavirus types 16 and 18. Cancer Epidemiol. Biomark. Prev. 2007, 16, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.F.; Schiffman, M.; Koutsky, L.A.; Hughes, J.P.; Winer, R.L.; Mao, C.; Hulbert, A.; Lee, S.K.; Shen, Z.; Kiviat, N.B. Lineages of oncogenic human papillomavirus types other than type 16 and 18 and risk for cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 2014, 106, dju270. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Yeager, M.; Cullen, M.; Boland, J.F.; Chen, Z.; Wentzensen, N.; Zhang, X.; Yu, K.; Yang, Q.; Mitchell, J.; et al. HPV16 Sublineage Associations With Histology-Specific Cancer Risk Using HPV Whole-Genome Sequences in 3200 Women. J. Natl. Cancer Inst. 2016, 108, djw100. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.A.; Gheit, T.; Franceschi, S.; Tommasino, M.; Clifford, G.M. Human Papillomavirus 18 Genetic Variation and Cervical Cancer Risk Worldwide. J. Virol. 2015, 89, 10680–10687. [Google Scholar] [CrossRef] [PubMed]
- Cullen, M.; Boland, J.F.; Schiffman, M.; Zhang, X.; Wentzensen, N.; Yang, Q.; Chen, Z.; Yu, K.; Mitchell, J.; Roberson, D.; et al. Deep sequencing of HPV16 genomes: A new high-throughput tool for exploring the carcinogenicity and natural history of HPV16 infection. Papillomavirus Res. 2015, 1, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Yeager, M.; Yu, K.; Clifford, G.M.; Xiao, Y.; Zhu, B.; Cullen, M.; Boland, J.F.; Wentzensen, N.; Nelson, C.W.; et al. HPV16 E7 Genetic Conservation Is Critical to Carcinogenesis. Cell 2017, 170, 1164–1174. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.F.; Schiffman, M.; Koutsky, L.A.; Hughes, J.P.; Hulbert, A.; Shen, Z.; Galloway, D.A.; Kiviat, N.B. Variant-specific persistence of infections with human papillomavirus Types 31, 33, 45, 56 and 58 and risk of cervical intraepithelial neoplasia. Int. J. Cancer 2016, 139, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Van Belkum, A.; Juffermans, L.; Schrauwen, L.; van Doornum, G.; Burger, M.; Quint, W. Genotyping human papillomavirus type 16 isolates from persistently infected promiscuous individuals and cervical neoplasia patients. J. Clin. Microbiol. 1995, 33, 2957–2962. [Google Scholar] [PubMed]
- Van der Weele, P.; Meijer, C.; King, A.J. Whole-Genome Sequencing and Variant Analysis of Hpv16 Infections. J. Virol. 2017, 92. [Google Scholar] [CrossRef]
- Chen, Z.; DeSalle, R.; Schiffman, M.; Herrero, R.; Burk, R.D. Evolutionary dynamics of variant genomes of human papillomavirus types 18, 45, and 97. J. Virol. 2009, 83, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- Lurchachaiwong, W.; Junyangdikul, P.; Termrungruanglert, W.; Payungporn, S.; Sampatanukul, P.; Tresukosol, D.; Niruthisard, S.; Trivijitsilp, P.; Karalak, A.; Swangvaree, S.; et al. Whole-genome sequence analysis of human papillomavirus type 18 from infected Thai women. Intervirology 2010, 53, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Schiffman, M.; Herrero, R.; DeSalle, R.; Anastos, K.; Segondy, M.; Sahasrabuddhe, V.V.; Gravitt, P.E.; Hsing, A.W.; Burk, R.D. Evolution and taxonomic classification of alphapapillomavirus 7 complete genomes: HPV18, HPV39, HPV45, HPV59, HPV68 and HPV70. PLoS ONE 2013, 8, e72565. [Google Scholar] [CrossRef] [PubMed]
- Apter, D.; Wheeler, C.M.; Paavonen, J.; Castellsague, X.; Garland, S.M.; Skinner, S.R.; Naud, P.; Salmeron, J.; Chow, S.N.; Kitchener, H.C.; et al. Efficacy of human papillomavirus 16 and 18 (HPV-16/18) AS04-adjuvanted vaccine against cervical infection and precancer in young women: Final event-driven analysis of the randomized, double-blind PATRICIA trial. Clin. Vaccine Immunol. 2015, 22, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Joura, E.A.; Giuliano, A.R.; Iversen, O.E.; Bouchard, C.; Mao, C.; Mehlsen, J.; Moreira, E.D., Jr.; Ngan, Y.; Petersen, L.K.; Lazcano-Ponce, E.; et al. A 9-valent HPV vaccine against infection and intraepithelial neoplasia in women. N. Engl. J. Med. 2015, 372, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Van den Broek, I.V.; Hoebe, C.J.; van Bergen, J.E.; Brouwers, E.E.; de Feijter, E.M.; Fennema, J.S.; Gotz, H.M.; Koekenbier, R.H.; van Ravesteijn, S.M.; de Coul, E.L. Evaluation design of a systematic, selective, internet-based, Chlamydia screening implementation in The Netherlands, 2008–2010: Implications of first results for the analysis. BMC Infect. Dis. 2010, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Van den Broek, I.V.; van Bergen, J.E.; Brouwers, E.E.; Fennema, J.S.; Gotz, H.M.; Hoebe, C.J.; Koekenbier, R.H.; Kretzschmar, M.; Over, E.A.; Schmid, B.V.; et al. Effectiveness of yearly, register based screening for chlamydia in The Netherlands: Controlled trial with randomised stepped wedge implementation. BMJ 2012, 345, e4316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mollers, M.; Boot Hein, J.; Vriend Henrike, J.; King Audrey, J.; van den Broek Ingrid, V.F.; van Bergen Jan, E.A.; Brink Antoinette, A.T.; Wolffs Petra, F.G.; Hoebe Christian, J.P.; Meijer Chris, J.L.; et al. Prevalence, incidence and persistence of genital HPV infections in a large cohort of sexually active young women in The Netherlands. Vaccine 2013, 31, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Woestenberg, P.J.; van Oeffelen, A.A.; Stirbu-Wagner, I.; van Benthem, B.H.; van Bergen, J.E.; van den Broek, I.V. Comparison of STI-related consultations among ethnic groups in The Netherlands: An epidemiologic study using electronic records from general practices. BMC Fam. Pract. 2015, 16, 70. [Google Scholar] [CrossRef] [PubMed]
- Kleter, B.; van Doorn, L.J.; Schrauwen, L.; Molijn, A.; Sastrowijoto, S.; ter Schegget, J.; Lindeman, J.; ter Harmsel, B.; Burger, M.; Quint, W. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 1999, 37, 2508–2517. [Google Scholar] [PubMed]
- Kleter, B.; van Doorn, L.J.; ter Schegget, J.; Schrauwen, L.; van Krimpen, K.; Burger, M.; ter Harmsel, B.; Quint, W. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am. J. Pathol. 1998, 153, 1731–1739. [Google Scholar] [CrossRef]
- Van der Weele, P.; van Logchem, E.; Wolffs, P.; van den Broek, I.; Feltkamp, M.; de Melker, H.; Meijer, C.J.; Boot, H.; King, A.J. Correlation between viral load, multiplicity of infection, and persistence of HPV16 and HPV18 infection in a Dutch cohort of young women. J. Clin. Virol. 2016, 83, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, S.L.; Basaras, M.; Arrese, E.; Hernaez, S.; Andia, D.; Esteban, V.; Garcia-Etxebarria, K.; Jugo, B.M.; Cisterna, R. Human papillomavirus (HPV) genotype 18 variants in patients with clinical manifestations of HPV related infections in Bilbao, Spain. Virol. J. 2012, 9, 258. [Google Scholar] [CrossRef] [PubMed]
- King, A.J.; Sonsma, J.A.; Vriend, H.J.; van der Sande, M.A.; Feltkamp, M.C.; Boot, H.J.; Koopmans, M.P.; Medical Microbiological, L.; Municipal Health, S. Genetic Diversity in the Major Capsid L1 Protein of HPV-16 and HPV-18 in The Netherlands. PLoS ONE 2016, 11, e0152782. [Google Scholar]
- Cole, S.T.; Danos, O. Nucleotide sequence and comparative analysis of the human papillomavirus type 18 genome. Phylogeny of papillomaviruses and repeated structure of the E6 and E7 gene products. J. Mol. Biol. 1987, 193, 599–608. [Google Scholar] [CrossRef]
- Van der Weele, P. National Institute for Public Health and the Environment (RIVM); Illumina Data of a HPV18 Apparent Reinfection Event; Centre for Infectious Disease Control: Bilthoven, The Netherlands, 2016. [Google Scholar]
- Siqueira, J.D.; Alves, B.M.; Prellwitz, I.M.; Furtado, C.; Meyrelles, A.R.; Machado, E.S.; Seuanez, H.N.; Soares, M.A.; Soares, E.A. Identification of novel human papillomavirus lineages and sublineages in HIV/HPV-coinfected pregnant women by next-generation sequencing. Virology 2016, 493, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.R.; Siqueira, J.D.; Finger-Jardim, F.; Vieira, V.C.; Silva, R.L.; Goncalves, C.V.; Soares, E.A.; Martinez, A.M.B.; Soares, M.A. Characterisation of complete high- and low-risk human papillomavirus genomes isolated from cervical specimens in southern Brazil. Memórias do Instituto Oswaldo Cruz 2017, 112, 728–731. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.F.; Kiviat, N.B.; Hildesheim, A.; Galloway, D.A.; Wheeler, C.M.; Ho, J.; Koutsky, L.A. Human papillomavirus type 16 and 18 variants: Race-related distribution and persistence. J. Natl. Cancer Inst. 2006, 98, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Lizano, M.; de la Cruz-Hernandez, E.; Carrillo-Garcia, A.; Garcia-Carranca, A.; Ponce de Leon-Rosales, S.; Duenas-Gonzalez, A.; Hernandez-Hernandez, D.M.; Mohar, A. Distribution of HPV16 and 18 intratypic variants in normal cytology, intraepithelial lesions, and cervical cancer in a Mexican population. Gynecol. Oncol. 2006, 102, 230–235. [Google Scholar] [CrossRef] [PubMed]

| General Subset Statistics | Infections (n Sequenced/n Total) | |
|---|---|---|
| Persistent | 25/66 | |
| Clearing | 26/79 | |
| Age | Median years (95% CI) | |
| Persistent | Clearing | |
| Total dataset | 24 (23–25) | 25 (23–26) |
| Sequenced subset | 24 (21–25) | 23 (20–26) |
| Unpaired t-test | p = 0.28 | p = 0.89 |
| C. trachomatis status | C. trachomatis positive (n)/Total (n) | |
| Persistent | Clearing | |
| Total dataset | 6/66 | 8/79 |
| Sequenced subset | 3/25 | 3/26 |
| Fisher’s exact test | p = 0.70 | p = 1 |
| European | European (n)/Total (n) | |
| Persistent | Clearing | |
| Total dataset | 51/66 | 55/79 |
| Sequenced subset | 18/25 | 19/26 |
| Fisher’s exact test | p = 0.59 | p = 0.81 |
| Mixed | Mixed (n)/Total (n) | |
| Persistent | Clearing | |
| Total dataset | 8/66 | 12/79 |
| Sequenced subset | 4/25 | 4/26 |
| Fisher’s exact test | p = 0.73 | p = 1.0 |
| Asian | Asian (n)/Total (n) | |
| Persistent | Clearing | |
| Total dataset | 6/66 | 8/79 |
| Sequenced subset | 3/25 | 2/26 |
| Fisher’s exact test | p = 0.70 | p = 1.0 |
| African | African (n)/Total (n) | |
| Persistent | Clearing | |
| Total dataset | 0/66 | 3/79 |
| Sequenced subset | 0/25 | 1/26 |
| Fisher’s exact test | p = 1.0 | p = 1.0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weele, P.V.d.; Meijer, C.J.L.M.; King, A.J. High Whole-Genome Sequence Diversity of Human Papillomavirus Type 18 Isolates. Viruses 2018, 10, 68. https://doi.org/10.3390/v10020068
Weele PVd, Meijer CJLM, King AJ. High Whole-Genome Sequence Diversity of Human Papillomavirus Type 18 Isolates. Viruses. 2018; 10(2):68. https://doi.org/10.3390/v10020068
Chicago/Turabian StyleWeele, Pascal Van der, Chris J.L.M. Meijer, and Audrey J. King. 2018. "High Whole-Genome Sequence Diversity of Human Papillomavirus Type 18 Isolates" Viruses 10, no. 2: 68. https://doi.org/10.3390/v10020068




