Proteomics Computational Analyses Suggest that the Envelope Glycoproteins of Segmented Jingmen Flavi-Like Viruses Are Class II Viral Fusion Proteins (β-Penetrenes) with Mucin-Like Domains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequences
2.2. Proteomics Computational Methods
3. Results
3.1. Sequence Similarities between Flavivirus and Flavi-Like Virus Proteins
3.2. Sequence Similarities of Jingmen Flavi-Like Virus Glycoproteins
3.3. Jingmen Flavi-Like Virus Glycoproteins Contain Predicted Mucin-Like Domains
3.4. Jingmen Flavi-Like Virus Glycoproteins Have Structure Features of Class II Viral Fusion Proteins
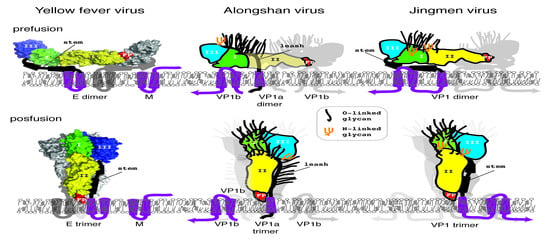
3.5. Jingmen Flavi-Like Virus Glycoprotein Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef]
- Gould, E.A.; de Lamballerie, X.; Zanotto, P.M.; Holmes, E.C. Origins, evolution, and vector/host coadaptations within the genus Flavivirus. Adv. Virus Res. 2003, 59, 277–314. [Google Scholar] [CrossRef]
- Cook, S.; Moureau, G.; Kitchen, A.; Gould, E.A.; de Lamballerie, X.; Holmes, E.C.; Harbach, R.E. Molecular evolution of the insect-specific flaviviruses. J. Gen. Virol. 2012, 93, 223–234. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Lin, X.D.; Vasilakis, N.; Tian, J.H.; Li, C.X.; Chen, L.J.; Eastwood, G.; Diao, X.N.; Chen, M.H.; Chen, X.; et al. Divergent Viruses Discovered in Arthropods and Vertebrates Revise the Evolutionary History of the Flaviviridae and Related Viruses. J. Virol. 2016, 90, 659–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bekal, S.; Domier, L.L.; Gonfa, B.; McCoppin, N.K.; Lambert, K.N.; Bhalerao, K. A novel flavivirus in the soybean cyst nematode. J. Gen. Virol. 2014, 95, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, G.; Tomita, R.; Kobayashi, K.; Sekine, K.T. Prevalence and genetic diversity of an unusual virus associated with Kobu-sho disease of gentian in Japan. J. Gen. Virol. 2013, 94, 2360–2365. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.C.; Shi, M.; Tian, J.H.; Lin, X.D.; Gao, D.Y.; He, J.R.; Wang, J.B.; Li, C.X.; Kang, Y.J.; Yu, B.; et al. A tick-borne segmented RNA virus contains genome segments derived from unsegmented viral ancestors. Proc. Natl. Acad. Sci. USA 2014, 111, 6744–6749. [Google Scholar] [CrossRef] [Green Version]
- Webster, C.L.; Longdon, B.; Lewis, S.H.; Obbard, D.J. Twenty-Five New Viruses Associated with the Drosophilidae (Diptera). Evol. Bioinform. Online 2016, 12, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Matsumura, E.E.; Nerva, L.; Nigg, J.C.; Falk, B.W.; Nouri, S. Complete Genome Sequence of the Largest Known Flavi-Like Virus, Diaphorina citri flavi-like virus, a Novel Virus of the Asian Citrus Psyllid, Diaphorina citri. Genome Announc. 2016, 4, e00946-16. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, M.; Sela, N.; Ng, J.; Casteel, C.L.; Peng, H.C.; Bekal, S.; Girke, T.; Ghanim, M.; Kaloshian, I. A novel virus from Macrosiphum euphorbiae with similarities to members of the family Flaviviridae. J. Gen. Virol. 2016, 97, 1261–1271. [Google Scholar] [CrossRef]
- Dincer, E.; Hacioglu, S.; Kar, S.; Emanet, N.; Brinkmann, A.; Nitsche, A.; Ozkul, A.; Linton, Y.M.; Ergunay, K. Survey and Characterization of Jingmen Tick Virus Variants. Viruses 2019, 11, 1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandegrift, K.J.; Kapoor, A. The Ecology of New Constituents of the Tick Virome and Their Relevance to Public Health. Viruses 2019, 11, 529. [Google Scholar] [CrossRef] [Green Version]
- Temmam, S.; Bigot, T.; Chretien, D.; Gondard, M.; Perot, P.; Pommelet, V.; Dufour, E.; Petres, S.; Devillers, E.; Hoem, T.; et al. Insights into the Host Range, Genetic Diversity, and Geographical Distribution of Jingmenviruses. mSphere 2019, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.D.; Wang, B.; Wei, F.; Han, S.Z.; Zhang, L.; Yang, Z.T.; Yan, Y.; Lv, X.L.; Li, L.; Wang, S.C.; et al. A New Segmented Virus Associated with Human Febrile Illness in China. N. Engl. J. Med. 2019, 380, 2116–2125. [Google Scholar] [CrossRef] [PubMed]
- Jia, N.; Liu, H.B.; Ni, X.B.; Bell-Sakyi, L.; Zheng, Y.C.; Song, J.L.; Li, J.; Jiang, B.G.; Wang, Q.; Sun, Y.; et al. Emergence of human infection with Jingmen tick virus in China: A retrospective study. EBioMedicine 2019, 43, 317–324. [Google Scholar] [CrossRef] [Green Version]
- Emmerich, P.; Jakupi, X.; von Possel, R.; Berisha, L.; Halili, B.; Gunther, S.; Cadar, D.; Ahmeti, S.; Schmidt-Chanasit, J. Viral metagenomics, genetic and evolutionary characteristics of Crimean-Congo hemorrhagic fever orthonairovirus in humans, Kosovo. Infect. Genet. Evol. 2018, 65, 6–11. [Google Scholar] [CrossRef]
- Sameroff, S.; Tokarz, R.; Charles, R.A.; Jain, K.; Oleynik, A.; Che, X.; Georges, K.; Carrington, C.V.; Lipkin, W.I.; Oura, C. Viral Diversity of Tick Species Parasitizing Cattle and Dogs in Trinidad and Tobago. Sci. Rep. 2019, 9, 10421. [Google Scholar] [CrossRef] [Green Version]
- Villa, E.C.; Maruyama, S.R.; de Miranda-Santos, I.K.F.; Palacios, G.; Ladner, J.T. Complete Coding Genome Sequence for Mogiana Tick Virus, a Jingmenvirus Isolated from Ticks in Brazil. Genome Announc. 2017, 5, e00232-17. [Google Scholar] [CrossRef] [Green Version]
- Pascoal, J.O.; Siqueira, S.M.; Maia, R.D.C.; Juan Szabo, M.P.; Yokosawa, J. Detection and molecular characterization of Mogiana tick virus (MGTV) in Rhipicephalus microplus collected from cattle in a savannah area, Uberlandia, Brazil. Ticks Tick-Borne Dis. 2019, 10, 162–165. [Google Scholar] [CrossRef]
- Ternovoi, V.A.; Protopopova, E.V.; Shvalov, A.N.; Kartashov, M.Y.; Bayandin, R.B.; Yakovlev, S.A.; Zakharov, K.S.; Nikiforov, A.K.; Konovalova, S.N.; Loktev, V.B.; et al. Complete Coding Genome Sequence for Novel Multicomponent Kindia Tick Virus Isolated from Ticks Collected in Guinea. Microbiol. Resour. Announc. submitted.
- Shen, S.; Moming, A.; Luo, T.; Chang, C.; Fang, Y.; Wang, J.; Kou, C.; Wang, C.; Su, Z.; Zhang, Y.; et al. Viromes of Hyalomma asiaticum, Hyalomma detritum and Dermacentor nuttalli ticks from Xinjiang Uygur Autonomous Region, China. GenBank 2018. Unpublished. [Google Scholar]
- Ladner, J.T.; Wiley, M.R.; Beitzel, B.; Auguste, A.J.; Dupuis, A.P., 2nd; Lindquist, M.E.; Sibley, S.D.; Kota, K.P.; Fetterer, D.; Eastwood, G.; et al. A Multicomponent Animal Virus Isolated from Mosquitoes. Cell Host Microbe 2016, 20, 357–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuivanen, S.; Levanov, L.; Kareinen, L.; Sironen, T.; Jaaskelainen, A.J.; Plyusnin, I.; Zakham, F.; Emmerich, P.; Schmidt-Chanasit, J.; Hepojoki, J.; et al. Detection of novel tick-borne pathogen, Alongshan virus, in Ixodes ricinus ticks, south-eastern Finland, 2019. Eurosurveillance 2019, 24, 1900394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallaher, W.R. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell 1996, 85, 477–478. [Google Scholar] [CrossRef] [Green Version]
- Gallaher, W.R.; Ball, J.M.; Garry, R.F.; Griffin, M.C.; Montelaro, R.C. A general model for the transmembrane proteins of HIV and other retroviruses. Aids Res. Hum. Retrovir. 1989, 5, 431–440. [Google Scholar] [CrossRef]
- Gallaher, W.R.; DiSimone, C.; Buchmeier, M.J. The viral transmembrane superfamily: Possible divergence of Arenavirus and Filovirus glycoproteins from a common RNA virus ancestor. BMC Microbiol 2001, 1, 1. [Google Scholar] [CrossRef] [Green Version]
- Garry, C.E.; Garry, R.F. Proteomics computational analyses suggest that the carboxyl terminal glycoproteins of Bunyaviruses are class II viral fusion proteins (β-penetrenes). Theor. Biol. Med. Model. 2004, 1, 10. [Google Scholar] [CrossRef] [Green Version]
- Garry, C.E.; Garry, R.F. Proteomics computational analyses suggest that baculovirus GP64 superfamily proteins are class III penetrenes. Virol. J. 2008, 5, 28. [Google Scholar] [CrossRef] [Green Version]
- Garry, C.E.; Garry, R.F. Proteomics computational analyses suggest that the bornavirus glycoprotein is a class III viral fusion protein (gamma penetrene). Virol. J. 2009, 6, 145. [Google Scholar] [CrossRef] [Green Version]
- Pearson, W. Lalign. Available online: https://embnet.vital-it.ch/software/LALIGN_form.html (accessed on 6 February 2020).
- Huang, X.; Miller, W. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 1991, 12, 337–357. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yachdav, G.; Kloppmann, E.; Kajan, L.; Hecht, M.; Goldberg, T.; Hamp, T.; Honigschmid, P.; Schafferhans, A.; Roos, M.; Bernhofer, M.; et al. PredictProtein--an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014, 42, W337–W343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, K.; Stoffel, W. TMpred. Available online: https://embnet.vital-it.ch/software/TMPRED_form.html (accessed on 6 February 2020).
- Hofmann, K.; Stoffel, W. TMbase—A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 1993, 374, 166. [Google Scholar]
- Snider, C.; Jayasinghe, S.; Hristova, K.; White, S.H. MPEx: A tool for exploring membrane proteins. Protein Sci. A Publ. Protein Soc. 2009, 18, 2624–2628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, S.H.; Wimley, W.C. Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta 1998, 1376, 339–352. [Google Scholar] [CrossRef] [Green Version]
- Wimley, W.C.; White, S.H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996, 3, 842–848. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef] [Green Version]
- Hrobowski, Y.M.; Garry, R.F.; Michael, S.F. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J. 2005, 2, 49. [Google Scholar] [CrossRef] [Green Version]
- Lok, S.M.; Costin, J.M.; Hrobowski, Y.M.; Hoffmann, A.R.; Rowe, D.K.; Kukkaro, P.; Holdaway, H.; Chipman, P.; Fontaine, K.A.; Holbrook, M.R.; et al. Release of dengue virus genome induced by a peptide inhibitor. PLoS ONE 2012, 7, e50995. [Google Scholar] [CrossRef]
- Allison, S.L.; Stiasny, K.; Stadler, K.; Mandl, C.W.; Heinz, F.X. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 1999, 73, 5605–5612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koehler, J.W.; Smith, J.M.; Ripoll, D.R.; Spik, K.W.; Taylor, S.L.; Badger, C.V.; Grant, R.J.; Ogg, M.M.; Wallqvist, A.; Guttieri, M.C.; et al. A fusion-inhibiting peptide against Rift Valley fever virus inhibits multiple, diverse viruses. PLoS Negl. Trop. Dis. 2013, 7, e2430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, I.B.; Gavel, Y.; von Heijne, G. Amino acid distributions around O-linked glycosylation sites. Biochem. J. 1991, 275 Pt 2, 529–534. [Google Scholar] [CrossRef] [Green Version]
- Bagdonaite, I.; Wandall, H.H. Global aspects of viral glycosylation. Glycobiology 2018, 28, 443–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.E.; Gruenke, J.A.; White, J.M. Leash in the groove mechanism of membrane fusion. Nat. Struct. Biol. 2003, 10, 1048–1053. [Google Scholar] [CrossRef] [PubMed]
- Badani, H.; Garry, R.F.; Wimley, W.C. Peptide entry inhibitors of enveloped viruses: The importance of interfacial hydrophobicity. Biochim. Biophys. Acta 2014, 1838, 2180–2197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epand, R.M. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta 2003, 1614, 116–121. [Google Scholar] [CrossRef] [Green Version]
- Gibbons, D.L.; Erk, I.; Reilly, B.; Navaza, J.; Kielian, M.; Rey, F.A.; Lepault, J. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell 2003, 114, 573–583. [Google Scholar] [CrossRef] [Green Version]
- Backovic, M.; Jardetzky, T.S. Class III viral membrane fusion proteins. Adv. Exp. Med. Biol. 2011, 714, 91–101. [Google Scholar] [CrossRef]
- Harrison, S.C. Viral membrane fusion. Virology 2015, 479–480, 498–507. [Google Scholar] [CrossRef] [Green Version]
- White, J.M.; Delos, S.E.; Brecher, M.; Schornberg, K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 189–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Modis, Y. Relating structure to evolution in class II viral membrane fusion proteins. Curr. Opin. Virol. 2014, 5, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Rey, F.A.; Heinz, F.X.; Mandl, C.; Kunz, C.; Harrison, S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 1995, 375, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Lescar, J.; Roussel, A.; Wien, M.W.; Navaza, J.; Fuller, S.D.; Wengler, G.; Wengler, G.; Rey, F.A. The Fusion glycoprotein shell of Semliki Forest virus: An icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 2001, 105, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Stiasny, K.; Fritz, R.; Pangerl, K.; Heinz, F.X. Molecular mechanisms of flavivirus membrane fusion. Amino Acids 2011, 41, 1159–1163. [Google Scholar] [CrossRef]
- Nayak, V.; Dessau, M.; Kucera, K.; Anthony, K.; Ledizet, M.; Modis, Y. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J. Virol. 2009, 83, 4338–4344. [Google Scholar] [CrossRef] [Green Version]
- Gibbons, D.L.; Vaney, M.C.; Roussel, A.; Vigouroux, A.; Reilly, B.; Lepault, J.; Kielian, M.; Rey, F.A. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 2004, 427, 320–325. [Google Scholar] [CrossRef]
- Li, L.; Lok, S.M.; Yu, I.M.; Zhang, Y.; Kuhn, R.J.; Chen, J.; Rossmann, M.G. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science 2008, 319, 1830–1834. [Google Scholar] [CrossRef] [Green Version]
- Pierson, T.C.; Diamond, M.S. Degrees of maturity: The complex structure and biology of flaviviruses. Curr. Opin. Virol. 2012, 2, 168–175. [Google Scholar] [CrossRef] [Green Version]
- Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 1981, 289, 366–373. [Google Scholar] [CrossRef]
- Duquerroy, S.; Vigouroux, A.; Rottier, P.J.; Rey, F.A.; Bosch, B.J. Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology 2005, 335, 276–285. [Google Scholar] [CrossRef] [Green Version]
- Heldwein, E.E.; Lou, H.; Bender, F.C.; Cohen, G.H.; Eisenberg, R.J.; Harrison, S.C. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 2006, 313, 217–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roche, S.; Bressanelli, S.; Rey, F.A.; Gaudin, Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 2006, 313, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Roche, S.; Rey, F.A.; Gaudin, Y.; Bressanelli, S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 2007, 315, 843–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavie, M.; Hanoulle, X.; Dubuisson, J. Glycan Shielding and Modulation of Hepatitis C Virus Neutralizing Antibodies. Front. Immunol. 2018, 9, 910. [Google Scholar] [CrossRef]
- Wagh, K.; Kreider, E.F.; Li, Y.; Barbian, H.J.; Learn, G.H.; Giorgi, E.; Hraber, P.T.; Decker, T.G.; Smith, A.G.; Gondim, M.V.; et al. Completeness of HIV-1 Envelope Glycan Shield at Transmission Determines Neutralization Breadth. Cell Rep. 2018, 25, 893–908.e7. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, Y.; Raghwani, J.; Allen, J.D.; Seabright, G.E.; Li, S.; Moser, F.; Huiskonen, J.T.; Strecker, T.; Bowden, T.A.; Crispin, M. Structure of the Lassa virus glycan shield provides a model for immunological resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 7320–7325. [Google Scholar] [CrossRef] [Green Version]
- Idris, F.; Muharram, S.H.; Diah, S. Glycosylation of dengue virus glycoproteins and their interactions with carbohydrate receptors: Possible targets for antiviral therapy. Arch. Virol. 2016, 161, 1751–1760. [Google Scholar] [CrossRef]
- Mossenta, M.; Marchese, S.; Poggianella, M.; Slon Campos, J.L.; Burrone, O.R. Role of N-glycosylation on Zika virus E protein secretion, viral assembly and infectivity. Biochem. Biophys. Res. Commun. 2017, 492, 579–586. [Google Scholar] [CrossRef]
- Zhang, X.; Jia, R.; Shen, H.; Wang, M.; Yin, Z.; Cheng, A. Structures and Functions of the Envelope Glycoprotein in Flavivirus Infections. Viruses 2017, 9, 338. [Google Scholar] [CrossRef] [Green Version]
- Stewart-Jones, G.B.; Soto, C.; Lemmin, T.; Chuang, G.Y.; Druz, A.; Kong, R.; Thomas, P.V.; Wagh, K.; Zhou, T.; Behrens, A.J.; et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell 2016, 165, 813–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.E.; Fusco, M.L.; Hessell, A.J.; Oswald, W.B.; Burton, D.R.; Saphire, E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 2008, 454, 177–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommerstein, R.; Flatz, L.; Remy, M.M.; Malinge, P.; Magistrelli, G.; Fischer, N.; Sahin, M.; Bergthaler, A.; Igonet, S.; Ter Meulen, J.; et al. Arenavirus Glycan Shield Promotes Neutralizing Antibody Evasion and Protracted Infection. PLoS Pathog. 2015, 11, e1005276. [Google Scholar] [CrossRef] [PubMed]
- Tran, E.E.; Simmons, J.A.; Bartesaghi, A.; Shoemaker, C.J.; Nelson, E.; White, J.M.; Subramaniam, S. Spatial localization of the Ebola virus glycoprotein mucin-like domain determined by cryo-electron tomography. J. Virol. 2014, 88, 10958–10962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peret, T.C.; Abed, Y.; Anderson, L.J.; Erdman, D.D.; Boivin, G. Sequence polymorphism of the predicted human metapneumovirus G glycoprotein. J. Gen. Virol. 2004, 85, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Pallesen, J.; Murin, C.D.; de Val, N.; Cottrell, C.A.; Hastie, K.M.; Turner, H.L.; Fusco, M.L.; Flyak, A.I.; Zeitlin, L.; Crowe, J.E., Jr.; et al. Structures of Ebola virus GP and sGP in complex with therapeutic antibodies. Nat. Microbiol. 2016, 1, 16128. [Google Scholar] [CrossRef]
- El Omari, K.; Iourin, O.; Harlos, K.; Grimes, J.M.; Stuart, D.I. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 2013, 3, 30–35. [Google Scholar] [CrossRef] [Green Version]
- Sabahi, A.; Uprichard, S.L.; Wimley, W.C.; Dash, S.; Garry, R.F. Minireview: Unexpected structural features of the hepatitis c virus envelope protein 2 ectodomain. J. Virol. 2014, 88, 10280–10288. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Giang, E.; Nieusma, T.; Kadam, R.U.; Cogburn, K.E.; Hua, Y.; Dai, X.; Stanfield, R.L.; Burton, D.R.; Ward, A.B.; et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science 2013, 342, 1090–1094. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.G.; Whidby, J.; Miller, M.T.; Scarborough, H.; Zatorski, A.V.; Cygan, A.; Price, A.A.; Yost, S.A.; Bohannon, C.D.; Jacob, J.; et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 2014, 509, 381–384. [Google Scholar] [CrossRef] [Green Version]







| Virus | Segment 1 | Segment 2 | Segment 3 | Segment 4 |
|---|---|---|---|---|
| Alongshan virus | MN107156.1 | MN107154.1 | MN107155.1 | MN107153.1 |
| Yanggou tick virus | MH688529.1 | MH688530.1 | MH688531.1 | MH688532.1 |
| Jingmen tick virus | MN025512.1 | MN025513.1 | MN025514.1 | MN025515.1 |
| Mogiana tick virus | NC_034222.1 | KY523073.1 | NC_034223.1 | KY523074.1 |
| Wuhan cricket virus | KR902709.1 | KR902710.1 | NC_028395.1 | NC_028402.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garry, C.E.; Garry, R.F. Proteomics Computational Analyses Suggest that the Envelope Glycoproteins of Segmented Jingmen Flavi-Like Viruses Are Class II Viral Fusion Proteins (β-Penetrenes) with Mucin-Like Domains. Viruses 2020, 12, 260. https://doi.org/10.3390/v12030260
Garry CE, Garry RF. Proteomics Computational Analyses Suggest that the Envelope Glycoproteins of Segmented Jingmen Flavi-Like Viruses Are Class II Viral Fusion Proteins (β-Penetrenes) with Mucin-Like Domains. Viruses. 2020; 12(3):260. https://doi.org/10.3390/v12030260
Chicago/Turabian StyleGarry, Courtney E., and Robert F. Garry. 2020. "Proteomics Computational Analyses Suggest that the Envelope Glycoproteins of Segmented Jingmen Flavi-Like Viruses Are Class II Viral Fusion Proteins (β-Penetrenes) with Mucin-Like Domains" Viruses 12, no. 3: 260. https://doi.org/10.3390/v12030260