Retroviral Integration Site Selection
Abstract
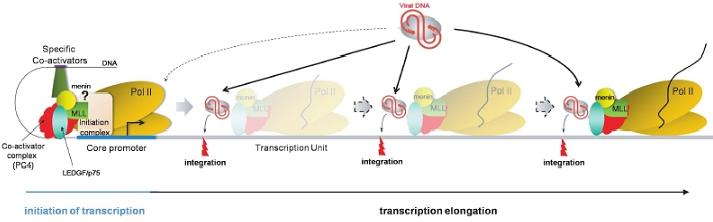
:1. Introduction

2. Integration targeting in vitro
3. Integration targeting in vivo
| Retroviridae genera | Specimen a | Host cell type b | Approx. Nb. of sites investigated c | References |
|---|---|---|---|---|
| lentiviruses | HIV-1 | human | 59869 | [23,36,59-76] |
| other | 2421 | [37,77-81] | ||
| HIV-2 | human | 202 | [82] | |
| SIV | human | 148 | [83] | |
| simian | 501 | [84] | ||
| EIAV | human | 1241 | [69,81] | |
| other | 70 | [81] | ||
| FIV | human | 226 | [85] | |
| alpharetroviruses | ASLV | human | 695 | [62,86] |
| avian | 658 | [77] | ||
| betaretroviruses | MMTV | human | 298 | [87] |
| murine | 170 | [87] | ||
| gammaretroviruses | MLV | human | 4005 | [60,66,70,73,88] |
| murine | 189 | [37] | ||
| other | 953 | [78,79,84] | ||
| MSCV | murine | 259 | [89] | |
| PERV | human | 1962 | [90,91] | |
| XMRV | human | 472 | [92] | |
| deltaretroviruses | HTLV-I | human | 1235 | [93-95] |
| epsilonretroviruses | Not investigated | |||
| spumaviruses | FV | human | 3457 | [65,70] |
| other | 263 | [78] | ||
| Endogenous retroviruses | HERV-K | human | 1565 | [88] |
| Retroviridae genera | in Transcription Unitsa | ± 2kb Transcription Start Sitesa | ± 2 kb CpG Islandsa |
|---|---|---|---|
| Lentiviruses | +/++ | 0 | -/0 |
| Alpharetroviruses | + | 0 | + |
| Betaretroviruses | 0 | 0 | 0 |
| Gammaretroviruses | + | ++ | ++ |
| Deltaretroviruses | + | + | + |
| Epsilonretroviruses | NA | NA | NA |
| Spumaviruses | 0 | ++ | ++ |
| HERV-class II | + | ++ | ++ |
3.1. The chromatin accessibility model
3.2. The cell cycle model
3.3. The tethering protein model
3.3.3. Epigenetic modifications and integration targeting
4. Conclusions

Acknowledgments
References
- Peterlin, B.M.; Trono, D. Hide, shield and strike back: how HIV-infected cells avoid immune eradication. Nat. Rev. Immunol. 2003, 3, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Goff, S.P. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 2007, 5, 253–263. [Google Scholar] [CrossRef]
- Suzuki, Y.; Craigie, R. The road to chromatin - nuclear entry of retroviruses. Nat. Rev. Microbiol. 2007, 5, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Flint, S.J.; Lynn W. Enquist, L.W.; Racaniello, V.R.; Skalka, A.M. Principles of Virology, 3rd edAmerican Society for Microbiology Press: Washington, DC, USA, 2009. [Google Scholar]
- Smith, J.A.; Daniel, R. Following the path of the virus: the exploitation of host DNA repair mechanisms by retroviruses. ACS Chem. Biol. 2006, 1, 217–226. [Google Scholar] [CrossRef]
- Vandegraaff, N.; Engelman, A. Molecular mechanisms of HIV integration and therapeutic intervention. Expert Rev. Mol. Med. 2007, 9, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Delelis, O.; Carayon, K.; Saib, A.; Deprez, E.; Mouscadet, J.F. Integrase and integration: biochemical activities of HIV-1 integrase. Retrovirology 2008, 5, 114. [Google Scholar] [CrossRef] [PubMed]
- Jaskolski, M.; Alexandratos, J.N.; Bujacz, G.; Wlodawer, A. Piecing together the structure of retroviral integrase, an important target in AIDS therapy. FEBS J. 2009, 276, 2926–2946. [Google Scholar] [CrossRef] [PubMed]
- Ceccherini-Silberstein, F.; Malet, I.; D'Arrigo, R.; Antinori, A.; Marcelin, A.G.; Perno, C.F. Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev. 2009, 11, 17–29. [Google Scholar] [PubMed]
- McKee, C.J.; Kessl, J.J.; Shkriabai, N.; Dar, M.J.; Engelman, A.; Kvaratskhelia, M. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J. Biol. Chem. 2008, 283, 31802–31812. [Google Scholar] [CrossRef] [PubMed]
- Lesbats, P.; Metifiot, M.; Calmels, C.; Baranova, S.; Nevinsky, G.; Andreola, M.L.; Parissi, V. In vitro initial attachment of HIV-1 integrase to viral ends: control of the DNA specific interaction by the oligomerization state. Nucleic Acids Res. 2008, 36, 7043–7058. [Google Scholar] [CrossRef] [PubMed]
- Hare, S.; Di Nunzio, F.; Labeja, A.; Wang, J.; Engelman, A.; Cherepanov, P. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 2009, 5, e1000515:1–e1000515:15. [Google Scholar]
- Michel, F.; Crucifix, C.; Granger, F.; Eiler, S.; Mouscadet, J.F.; Korolev, S.; Agapkina, J.; Ziganshin, R.; Gottikh, M.; Nazabal, A.; Emiliani, S.; Benarous, R.; Moras, D.; Schultz, P.; Ruff, M. Structural basis for HIV-1 DNA integration in the human genome, role of the LEDGF/P75 cofactor. Embo J. 2009, 28, 980–991. [Google Scholar] [CrossRef] [PubMed]
- Hare, S.; Cherepanov, P.; Wang, J. Application of general formulas for the correction of a lattice-translocation defect in crystals of a lentiviral integrase in complex with LEDGF. Acta Crystallogr. D. Biol. Crystallogr. 2009, 65, 966–973. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. HIV-1 gene expression: lessons from provirus and non-integrated DNA. Retrovirology 2004, 1, 13. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. The second chance story of HIV-1 DNA: Unintegrated? Not a problem! Retrovirology 2008, 5, 61. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.R.; Yu, D.; Biancotto, A.; Margolis, L.B.; Wu, Y. Measurement of human immunodeficiency virus type 1 preintegration transcription by using Rev-dependent Rev-CEM cells reveals a sizable transcribing DNA population comparable to that from proviral templates. J. Virol. 2009, 83, 8662–8673. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.A.; Ott, D.E.; Gorelick, R.J. Efficiency of human immunodeficiency virus type 1 postentry infection processes: evidence against disproportionate numbers of defective virions. J. Virol. 2007, 81, 4367–4370. [Google Scholar] [CrossRef] [PubMed]
- Albanese, A.; Arosio, D.; Terreni, M.; Cereseto, A. HIV-1 pre-integration complexes selectively target decondensed chromatin in the nuclear periphery. PLoS ONE 2008, 3, e2413:1–e2413:9. [Google Scholar] [CrossRef]
- Christ, F.; Thys, W.; De Rijck, J.; Gijsbers, R.; Albanese, A.; Arosio, D.; Emiliani, S.; Rain, J.C.; Benarous, R.; Cereseto, A.; Debyser, Z. Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 2008, 18, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Jordan, A.; Defechereux, P.; Verdin, E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. Embo J. 2001, 20, 1726–1738. [Google Scholar] [CrossRef] [PubMed]
- Jordan, A.; Bisgrove, D.; Verdin, E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. Embo J. 2003, 22, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Lewinski, M.K.; Bisgrove, D.; Shinn, P.; Chen, H.; Hoffmann, C.; Hannenhalli, S.; Verdin, E.; Berry, C.C.; Ecker, J.R.; Bushman, F.D. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 2005, 79, 6610–6619. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Lin, Y.B.; An, W.; Xu, J.; Yang, H.C.; O'Connell, K.; Dordai, D.; Boeke, J.D.; Siliciano, J.D.; Siliciano, R.F. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 2008, 4, 134–146. [Google Scholar] [CrossRef]
- Bushman, F.D. Lateral DNA Transfer: Mechanisms and Consequences. 2001; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA. [Google Scholar]
- Voigt, K.; Izsvak, Z.; Ivics, Z. Targeted gene insertion for molecular medicine. J. Mol. Med. 2008, 86, 1205–1219. [Google Scholar] [CrossRef] [PubMed]
- Nienhuis, A.W.; Dunbar, C.E.; Sorrentino, B.P. Genotoxicity of retroviral integration in hematopoietic cells. Mol. Ther. 2006, 13, 1031–1049. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.D. Retroviral integration and human gene therapy. J. Clin. Invest. 2007, 117, 2083–2086. [Google Scholar] [CrossRef] [PubMed]
- Pike-Overzet, K.; van der Burg, M.; Wagemaker, G.; van Dongen, J.J.; Staal, F.J. New insights and unresolved issues regarding insertional mutagenesis in X-linked SCID gene therapy. Mol. Ther. 2007, 15, 1910–1916. [Google Scholar] [CrossRef] [PubMed]
- Cavazzana-Calvo, M.; Fischer, A. Gene therapy for severe combined immunodeficiency: are we there yet? J. Clin. Invest. 2007, 117, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Kimmelman, J. The ethics of human gene transfer. Nat. Rev. Genet. 2008, 9, 239–244. [Google Scholar] [CrossRef]
- Ciuffi, A. Mechanisms governing lentivirus integration site selection. Curr. Gene Ther. 2008, 8, 419–429. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; von Kalle, C.; Schmidt, M.; Le Deist, F.; Wulffraat, N.; McIntyre, E.; Radford, I.; Villeval, J.L.; Fraser, C.C.; Cavazzana-Calvo, M.; Fischer, A. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003, 348, 255–256. [Google Scholar] [CrossRef] [PubMed]
- Hacein-Bey-Abina, S.; Garrigue, A.; Wang, G.P.; Soulier, J.; Lim, A.; Morillon, E.; Clappier, E.; Caccavelli, L.; Delabesse, E.; Beldjord, K.; Asnafi, V.; MacIntyre, E.; Dal Cortivo, L.; Radford, I.; Brousse, N.; Sigaux, F.; Moshous, D.; Hauer, J.; Borkhardt, A.; Belohradsky, B.H.; Wintergerst, U.; Velez, M.C.; Leiva, L.; Sorensen, R.; Wulffraat, N.; Blanche, S.; Bushman, F.D.; Fischer, A.; Cavazzana-Calvo, M. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008, 118, 3132–3142. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.P.; Garrigue, A.; Ciuffi, A.; Ronen, K.; Leipzig, J.; Berry, C.; Lagresle-Peyrou, C.; Benjelloun, F.; Hacein-Bey-Abina, S.; Fischer, A.; Cavazzana-Calvo, M.; Bushman, F.D. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008, 36, e49:1–e49:12. [Google Scholar]
- Wang, G.P.; Levine, B.L.; Binder, G.K.; Berry, C.C.; Malani, N.; McGarrity, G.; Tebas, P.; June, C.H.; Bushman, F.D. Analysis of Lentiviral Vector Integration in HIV+ Study Subjects Receiving Autologous Infusions of Gene Modified CD4+ T Cells. Mol. Ther. 2009, 17, 844–850. [Google Scholar] [CrossRef] [PubMed]
- Montini, E.; Cesana, D.; Schmidt, M.; Sanvito, F.; Bartholomae, C.C.; Ranzani, M.; Benedicenti, F.; Sergi, L.S.; Ambrosi, A.; Ponzoni, M.; Doglioni, C.; Di Serio, C.; von Kalle, C.; Naldini, L. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J. Clin. Invest. 2009, 119, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.D.; Fujiwara, T.; Craigie, R. Retroviral DNA integration directed by HIV integration protein in vitro. Science 1990, 249, 1555–1558. [Google Scholar] [PubMed]
- Chow, S.A. In vitro assays for activities of retroviral integrase. Methods 1997, 12, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Pursley, M.H.; Grandgenett, D.P. Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. J. Virol. 2002, 76, 3105–3113. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Grandgenett, D.P. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J. Virol. 2005, 79, 8208–8216. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.D.; Craigie, R. Sequence requirements for integration of Moloney murine leukemia virus DNA in vitro. J. Virol. 1990, 64, 5645–5648. [Google Scholar] [PubMed]
- Leavitt, A.D.; Rose, R.B.; Varmus, H.E. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J. Virol. 1992, 66, 2359–2368. [Google Scholar] [PubMed]
- Pryciak, P.M.; Muller, H.P.; Varmus, H.E. Simian virus 40 minichromosomes as targets for retroviral integration in vivo. Proc. Natl. Acad. Sci. U. S. A. 1992, 89, 9237–9241. [Google Scholar] [CrossRef] [PubMed]
- Pryciak, P.M.; Sil, A.; Varmus, H.E. Retroviral integration into minichromosomes in vitro. Embo J. 1992, 11, 291–303. [Google Scholar] [PubMed]
- Pryciak, P.M.; Varmus, H.E. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell 1992, 69, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Bor, Y.C.; Bushman, F.D.; Orgel, L.E. In vitro integration of human immunodeficiency virus type 1 cDNA into targets containing protein-induced bends. Proc. Natl. Acad. Sci. USA 1995, 92, 10334–10338. [Google Scholar] [CrossRef]
- Stevens, S.W.; Griffith, J.D. Sequence analysis of the human DNA flanking sites of human immunodeficiency virus type 1 integration. J. Virol. 1996, 70, 6459–6462. [Google Scholar] [PubMed]
- Bor, Y.C.; Miller, M.D.; Bushman, F.D.; Orgel, L.E. Target-sequence preferences of HIV-1 integration complexes in vitro. Virology 1996, 222, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.P.; Varmus, H.E. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. Embo J. 1994, 13, 4704–4714. [Google Scholar] [PubMed]
- Pruss, D.; Bushman, F.D.; Wolffe, A.P. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. USA 1994, 91, 5913–5917. [Google Scholar] [CrossRef]
- Pruss, D.; Reeves, R.; Bushman, F.D.; Wolffe, A.P. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biol. Chem. 1994, 269, 25031–25041. [Google Scholar] [PubMed]
- Botbol, Y.; Raghavendra, N.K.; Rahman, S.; Engelman, A.; Lavigne, M. Chromatinized templates reveal the requirement for the LEDGF/p75 PWWP domain during HIV-1 integration in vitro. Nucleic Acids Res. 2008, 36, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.D. Tethering human immunodeficiency virus 1 integrase to a DNA site directs integration to nearby sequences. Proc. Natl. Acad. Sci. USA 1994, 91, 9233–9237. [Google Scholar] [CrossRef]
- Goulaouic, H.; Chow, S.A. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J. Virol. 1996, 70, 37–46. [Google Scholar] [PubMed]
- Tan, W.; Zhu, K.; Segal, D.J.; Barbas 3rd, C.F.; Chow, S.A. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J. Virol. 2004, 78, 1301–1313. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.; Lewinski, M.; Ciuffi, A.; Barr, S.; Leipzig, J.; Hannenhalli, S.; Hoffmann, C. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005, 3, 848–858. [Google Scholar] [CrossRef]
- Ciuffi, A.; Ronen, K.; Brady, T.; Malani, N.; Wang, G.; Berry, C.C.; Bushman, F.D. Methods for integration site distribution analyses in animal cell genomes. Methods 2009, 47, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Schroder, A.R.; Shinn, P.; Chen, H.; Berry, C.; Ecker, J.R.; Bushman, F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 2002, 110, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, Y.; Crise, B.; Burgess, S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science 2003, 300, 1749–1751. [Google Scholar] [CrossRef] [PubMed]
- Craigie, R.; Fujiwara, T.; Bushman, F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 1990, 62, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.S.; Beitzel, B.F.; Schroder, A.R.; Shinn, P.; Chen, H.; Berry, C.C.; Ecker, J.R.; Bushman, F.D. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004, 2, e234:1127–e234:1137. [Google Scholar] [CrossRef] [Green Version]
- Ciuffi, A.; Llano, M.; Poeschla, E.; Hoffmann, C.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005, 11, 1287–1289. [Google Scholar] [CrossRef] [PubMed]
- Ciuffi, A.; Mitchell, R.S.; Hoffmann, C.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F.D. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Mol. Ther. 2006, 13, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Trobridge, G.D.; Miller, D.G.; Jacobs, M.A.; Allen, J.M.; Kiem, H.P.; Kaul, R.; Russell, D.W. Foamy virus vector integration sites in normal human cells. Proc. Natl. Acad. Sci. USA 2006, 103, 1498–1503. [Google Scholar] [CrossRef]
- Lewinski, M.K.; Yamashita, M.; Emerman, M.; Ciuffi, A.; Marshall, H.; Crawford, G.; Collins, F.; Shinn, P.; Leipzig, J.; Hannenhalli, S.; Berry, C.C.; Ecker, J.R.; Bushman, F.D. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006, 2, e60:611–e60:622. [Google Scholar] [CrossRef]
- Barr, S.D.; Ciuffi, A.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F.D. HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry. Mol. Ther. 2006, 14, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.L.; Humeau, L.M.; Boyer, J.; MacGregor, R.R.; Rebello, T.; Lu, X.; Binder, G.K.; Slepushkin, V.; Lemiale, F.; Mascola, J.R.; Bushman, F.D.; Dropulic, B.; June, C.H. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc. Natl. Acad. Sci. USA 2006, 103, 17372–17377. [Google Scholar] [CrossRef]
- Hacker, C.V.; Vink, C.A.; Wardell, T.W.; Lee, S.; Treasure, P.; Kingsman, S.M.; Mitrophanous, K.A.; Miskin, J.E. The integration profile of EIAV-based vectors. Mol. Ther. 2006, 14, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Nowrouzi, A.; Dittrich, M.; Klanke, C.; Heinkelein, M.; Rammling, M.; Dandekar, T.; von Kalle, C.; Rethwilm, A. Genome-wide mapping of foamy virus vector integrations into a human cell line. J. Gen. Virol. 2006, 87, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, Y.; Liang, T.; Sinsheimer, J.S.; Chow, S.A. A high-throughput method for cloning and sequencing human immunodeficiency virus type 1 integration sites. J. Virol. 2006, 80, 11313–11321. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.P.; Ciuffi, A.; Leipzig, J.; Berry, C.C.; Bushman, F.D. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007, 17, 1186–1194. [Google Scholar] [CrossRef] [PubMed]
- Felice, B.; Cattoglio, C.; Cittaro, D.; Testa, A.; Miccio, A.; Ferrari, G.; Luzi, L.; Recchia, A.; Mavilio, F. Transcription factor binding sites are genetic determinants of retroviral integration in the human genome. PLoS ONE 2009, 4, e4571:1–e4571:16. [Google Scholar] [CrossRef]
- Wellensiek, B.P.; Ramakrishnan, R.; Sundaravaradan, V.; Mehta, R.; Harris, D.T.; Ahmad, N. Differential HIV-1 integration targets more actively transcribed host genes in neonatal than adult blood mononuclear cells. Virology 2009, 385, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Vatakis, D.N.; Kim, S.; Kim, N.; Chow, S.A.; Zack, J.A. Human immunodeficiency virus integration efficiency and site selection in quiescent CD4+ T cells. J. Virol. 2009, 83, 6222–6233. [Google Scholar] [CrossRef] [PubMed]
- Brady, T.; Agosto, L.M.; Malani, N.; Berry, C.C.; O'Doherty, U.; Bushman, F. HIV integration site distributions in resting and activated CD4+ T cells infected in culture. Aids 2009, 23, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- Barr, S.D.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F.D. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J. Virol. 2005, 79, 12035–12044. [Google Scholar] [CrossRef] [PubMed]
- Beard, B.C.; Keyser, K.A.; Trobridge, G.D.; Peterson, L.J.; Miller, D.G.; Jacobs, M.; Kaul, R.; Kiem, H.P. Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, or foamy virus. Hum. Gene Ther. 2007, 18, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Beard, B.C.; Dickerson, D.; Beebe, K.; Gooch, C.; Fletcher, J.; Okbinoglu, T.; Miller, D.G.; Jacobs, M.A.; Kaul, R.; Kiem, H.P.; Trobridge, G.D. Comparison of HIV-derived Lentiviral and MLV-based Gammaretroviral Vector Integration Sites in Primate Repopulating Cells. Mol. Ther. 2007, 15, 1356–1365. [Google Scholar] [CrossRef] [PubMed]
- Shun, M.C.; Raghavendra, N.K.; Vandegraaff, N.; Daigle, J.E.; Hughes, S.; Kellam, P.; Cherepanov, P.; Engelman, A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007, 21, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Marshall, H.M.; Ronen, K.; Berry, C.; Llano, M.; Sutherland, H.; Saenz, D.; Bickmore, W.; Poeschla, E.; Bushman, F.D. Role of PSIP1/LEDGF/p75 in Lentiviral Infectivity and Integration Targeting. PLoS ONE 2007, 2, e1340:1–e1340:13. [Google Scholar]
- MacNeil, A.; Sankale, J.L.; Meloni, S.T.; Sarr, A.D.; Mboup, S.; Kanki, P. Genomic sites of human immunodeficiency virus type 2 (HIV-2) integration: similarities to HIV-1 in vitro and possible differences in vivo. J. Virol. 2006, 80, 7316–7321. [Google Scholar] [CrossRef] [PubMed]
- Crise, B.; Li, Y.; Yuan, C.; Morcock, D.R.; Whitby, D.; Munroe, D.J.; Arthur, L.O.; Wu, X. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J. Virol. 2005, 79, 12199–12204. [Google Scholar] [CrossRef] [PubMed]
- Hematti, P.; Hong, B.K.; Ferguson, C.; Adler, R.; Hanawa, H.; Sellers, S.; Holt, I.E.; Eckfeldt, C.E.; Sharma, Y.; Schmidt, M.; von Kalle, C.; Persons, D.A.; Billings, E.M.; Verfaillie, C.M.; Nienhuis, A.W.; Wolfsberg, T.G.; Dunbar, C.E.; Calmels, B. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004, 2, e423:1–e423:8. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.; Moressi, C.J.; Scheetz, T.E.; Xie, L.; Tran, D.T.; Casavant, T.L.; Ak, P.; Benham, C.J.; Davidson, B.L.; McCray Jr. Jr, P.B. Integration site choice of a feline immunodeficiency virus vector. J. Virol. 2006, 80, 8820–8823. [Google Scholar] [CrossRef] [PubMed]
- Narezkina, A.; Taganov, K.D.; Litwin, S.; Stoyanova, R.; Hayashi, J.; Seeger, C.; Skalka, A.M.; Katz, R.A. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 2004, 78, 11656–11663. [Google Scholar] [CrossRef] [PubMed]
- Faschinger, A.; Rouault, F.; Sollner, J.; Lukas, A.; Salmons, B.; Gunzburg, W.H.; Indik, S. Mouse Mammary Tumor Virus Integration Site Selection in Human and Mouse Genomes. J. Virol. 2008, 83, 1360–1367. [Google Scholar] [CrossRef]
- Brady, T.; Lee, Y.N.; Ronen, K.; Malani, N.; Berry, C.C.; Bieniasz, P.D.; Bushman, F.D. Integration target site selection by a resurrected human endogenous retrovirus. Genes Dev. 2009, 23, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Aker, M.; Tubb, J.; Miller, D.G.; Stamatoyannopoulos, G.; Emery, D.W. Integration bias of gammaretrovirus vectors following transduction and growth of primary mouse hematopoietic progenitor cells with and without selection. Mol. Ther. 2006, 14, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Moalic, Y.; Blanchard, Y.; Felix, H.; Jestin, A. Porcine endogenous retrovirus integration sites in the human genome: features in common with those of murine leukemia virus. J. Virol. 2006, 80, 10980–10988. [Google Scholar] [CrossRef] [PubMed]
- Moalic, Y.; Felix, H.; Takeuchi, Y.; Jestin, A.; Blanchard, Y. Genome areas with high gene density and CpG island neighborhood strongly attract porcine endogenous retrovirus for integration and favor the formation of hot spots. J. Virol. 2009, 83, 1920–1929. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, N.; Dong, B.; Boren, D.; Lee, S.A.; Das Gupta, J.; Gaughan, C.; Klein, E.A.; Lee, C.; Silverman, R.H.; Chow, S.A. Integration site preference of xenotropic murine leukemia virus-related virus, a new human retrovirus associated with prostate cancer. J. Virol. 2008, 82, 9964–9977. [Google Scholar] [CrossRef] [PubMed]
- Doi, K.; Wu, X.; Taniguchi, Y.; Yasunaga, J.; Satou, Y.; Okayama, A.; Nosaka, K.; Matsuoka, M. Preferential selection of human T-cell leukemia virus type I provirus integration sites in leukemic versus carrier states. Blood 2005, 106, 1048–1053. [Google Scholar] [CrossRef] [PubMed]
- Derse, D.; Crise, B.; Li, Y.; Princler, G.; Lum, N.; Stewart, C.; McGrath, C.F.; Hughes, S.H.; Munroe, D.J.; Wu, X. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J. Virol. 2007, 81, 6731–6741. [Google Scholar] [CrossRef] [PubMed]
- Meekings, K.N.; Leipzig, J.; Bushman, F.D.; Taylor, G.P.; Bangham, C.R. HTLV-1 Integration into Transcriptionally Active Genomic Regions Is Associated with Proviral Expression and with HAM/TSP. PLoS Pathog. 2008, 4, e1000027:1–e1000027:10. [Google Scholar] [CrossRef]
- Carteau, S.; Hoffmann, C.; Bushman, F. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J. Virol. 1998, 72, 4005–4014. [Google Scholar] [PubMed]
- Holman, A.G.; Coffin, J.M. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc. Natl. Acad. Sci. USA 2005, 102, 6103–6107. [Google Scholar] [CrossRef]
- Berry, C.; Hannenhalli, S.; Leipzig, J.; Bushman, F.D. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2006, 2, e157:1450–e157:1462. [Google Scholar] [CrossRef]
- Shun, M.C.; Daigle, J.E.; Vandegraaff, N.; Engelman, A. Wild-type levels of human immunodeficiency virus type 1 infectivity in the absence of cellular emerin protein. J. Virol. 2007, 81, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Segal, E.; Fondufe-Mittendorf, Y.; Chen, L.; Thastrom, A.; Field, Y.; Moore, I.K.; Wang, J.P.; Widom, J. A genomic code for nucleosome positioning. Nature 2006, 442, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Crawford, G.E.; Holt, I.E.; Whittle, J.; Webb, B.D.; Tai, D.; Davis, S.; Margulies, E.H.; Chen, Y.; Bernat, J.A.; Ginsburg, D.; Zhou, D.; Luo, S.; Vasicek, T.J.; Daly, M.J.; Wolfsberg, T.G.; Collins, F.S. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res. 2006, 16, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Tobaly-Tapiero, J.; Bittoun, P.; Lehmann-Che, J.; Delelis, O.; Giron, M.L.; de The, H.; Saib, A. Chromatin tethering of incoming foamy virus by the structural Gag protein. Traffic 2008, 9, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.D. Integration site selection by lentiviruses: biology and possible control. Curr. Top. Microbiol. Immunol. 2002, 261, 165–177. [Google Scholar] [PubMed]
- Engelman, A. The roles of cellular factors in retroviral integration. Curr. Top. Microbiol. Immunol. 2003, 281, 209–238. [Google Scholar] [PubMed]
- Van Maele, B.; Debyser, Z. HIV-1 integration: an interplay between HIV-1 integrase, cellular and viral proteins. AIDS Rev. 2005, 7, 26–43. [Google Scholar] [PubMed]
- Van Maele, B.; Busschots, K.; Vandekerckhove, L.; Christ, F.; Debyser, Z. Cellular co-factors of HIV-1 integration. Trends Biochem. Sci. 2006, 31, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Al-Mawsawi, L.Q.; Neamati, N. Blocking interactions between HIV-1 integrase and cellular cofactors: an emerging anti-retroviral strategy. Trends Pharmacol. Sci. 2007, 28, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.D.; Malani, N.; Fernandes, J.; D'Orso, I.; Cagney, G.; Diamond, T.L.; Zhou, H.; Hazuda, D.J.; Espeseth, A.S.; Konig, R.; Bandyopadhyay, S.; Ideker, T.; Goff, S.P.; Krogan, N.J.; Frankel, A.D.; Young, J.A.; Chanda, S.K. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009, 5, e1000437:1–e1000437:12. [Google Scholar] [CrossRef]
- Ciuffi, A.; Bushman, F.D. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006, 22, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.; Cherepanov, P. The Lentiviral Integrase Binding Protein LEDGF/p75 and HIV-1 Replication. PLoS Pathog. 2008, 4, e1000046:1–e1000046:9. [Google Scholar] [CrossRef]
- Poeschla, E.M. Integrase, LEDGF/p75 and HIV replication. Cell. Mol. Life Sci. 2008, 65, 1403–1424. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, P.; Ambrosio, A.L.; Rahman, S.; Ellenberger, T.; Engelman, A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. USA 2005, 102, 17308–17313. [Google Scholar] [CrossRef]
- Ge, H.; Si, Y.; Roeder, R.G. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. Embo J. 1998, 17, 6723–6729. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, V.; Daniels, T.; Casiano, C.A. LEDGF/p75: a novel nuclear autoantigen at the crossroads of cell survival and apoptosis. Autoimmun. Rev. 2003, 2, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, T.; Singh, D.P.; Fatma, N. LEDGF, a survival factor, activates stress-related genes. Prog. Retin. Eye Res. 2002, 21, 341–358. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, H.G.; Newton, K.; Brownstein, D.G.; Holmes, M.C.; Kress, C.; Semple, C.A.; Bickmore, W.A. Disruption of Ledgf/Psip1 results in perinatal mortality and homeotic skeletal transformations. Mol. Cell. Biol. 2006, 26, 7201–7210. [Google Scholar] [CrossRef] [PubMed]
- Llano, M.; Saenz, D.T.; Meehan, A.; Wongthida, P.; Peretz, M.; Walker, W.H.; Teo, W.; Poeschla, E.M. An essential role for LEDGF/p75 in HIV integration. Science 2006, 314, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Vandekerckhove, L.; Christ, F.; Van Maele, B.; De Rijck, J.; Gijsbers, R.; Van den Haute, C.; Witvrouw, M.; Debyser, Z. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 2006, 80, 1886–1896. [Google Scholar] [CrossRef] [PubMed]
- De Rijck, J. and Debyser. KU Leuven, Leuven, Belgium.Personal communication. 2009. [Google Scholar]
- Desfarges, S.; Munoz, M.; Leleu, M.; Lefebvre, G.; Rougemont, J.; Xenarios, I.; Ciuffi, A. Analysis of LEDGF/p75-bound sequences by ChIP-Seq (to be submitted for publication). Genome Res.
- Ciuffi, A.; Diamond, T.L.; Hwang, Y.; Marshall, H.M.; Bushman, F.D. Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Hum. Gene Ther. 2006, 17, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Meehan, A.M.; Saenz, D.T.; Morrison, J.H.; Garcia-Rivera, J.A.; Peretz, M.; Llano, M.; Poeschla, E.M. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog. 2009, 5, e1000522:1–e1000522:18. [Google Scholar] [CrossRef]
- Silvers, R.; Smith, J.A.; Schowalter, M.; Litwin, S.; Liang, Z.; Geary, K.; Daniel, R. Modification of Integration Site Preferences of an HIV-1-based Vector by expression of a novel synthetic protein. Hum. Gene Ther. 2009. [Google Scholar]
- Gijsbers, R.; Ronen, K.; Vets, S.; Malani, N.; De Rijck, J.; McNeely, M.; Bushman, F.D.; Debyser, Z. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol. Ther. 2010, In Press. [Google Scholar]
- Studamire, B.; Goff, S.P. Host proteins interacting with the Moloney murine leukemia virus integrase: multiple transcriptional regulators and chromatin binding factors. Retrovirology 2008, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.E.P. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004, 306, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.E.P. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.; Cleary, M.L. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 2008, 14, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Roeder, R.G. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 1994, 78, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Roeder, R.G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996, 21, 327–335. [Google Scholar] [PubMed]
- Fukuda, A.; Nakadai, T.; Shimada, M.; Tsukui, T.; Matsumoto, M.; Nogi, Y.; Meisterernst, M.; Hisatake, K. Transcriptional coactivator PC4 stimulates promoter escape and facilitates transcriptional synergy by GAL4-VP16. Mol. Cell. Biol. 2004, 24, 6525–6535. [Google Scholar] [CrossRef] [PubMed]
- Calvo, O.; Manley, J.L. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. Embo J. 2005, 24, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Desfarges, S.; Ciuffi, A. Retroviral Integration Site Selection. Viruses 2010, 2, 111-130. https://doi.org/10.3390/v2010111
Desfarges S, Ciuffi A. Retroviral Integration Site Selection. Viruses. 2010; 2(1):111-130. https://doi.org/10.3390/v2010111
Chicago/Turabian StyleDesfarges, Sébastien, and Angela Ciuffi. 2010. "Retroviral Integration Site Selection" Viruses 2, no. 1: 111-130. https://doi.org/10.3390/v2010111