The Role of Interferon Antagonist, Non-Structural Proteins in the Pathogenesis and Emergence of Arboviruses
Abstract
:1. Introduction
2. Emergence
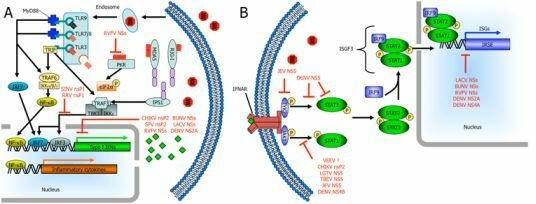
3. Interferon Responses to Viral Infection
| Family | Nonstructural Proteins | Direct IFN Antagonism | Host Transcriptional Shut off as Indirect Mechanism for IFN Antagonism | References |
|---|---|---|---|---|
| Genus | ||||
| Virus | ||||
| Bunyaviridae | ||||
| Orthobunyavirus | ||||
| La Crosse virus | NSs | -Inhibits RNA Pol II transcription by triggering degradation of RPB1 | [72] | |
| Bunyamwera virus | NSs | -Inhibits RNA Pol II transcription by blocking elongation | [84,85] | |
| Phlebovirus | ||||
| Rift Valley Fever virus | NSs | -Degrades PKR -Interacts with SAP30 to block activation of the IFN-β promoter | -Inhibits RNA Pol II transcription by preventing TFIIH assembly | [78,83,100] |
| Togaviridae | ||||
| Alphavirus | ||||
| Venezuelan Equine Encephalitis virus | nsP2? | -Blocks STAT1 phosphorylation | -Transcriptional shutoff by capsid protein | [17,111] |
| Chikungunya virus | nsP2 | -Blocks STAT1 nuclear import | -Transcriptional shutoff (mechanism unknown) | [113,115] |
| Sindbis virus | nsP2 | -Blocks NF-κB-dependent PRDII promoters? | -Downregulation of RNA Pol I and II-dependent transcription | [16,118] |
| Semliki Forrest virus | nsP2 | -Cleavage of transcription factors (suggested) | -Transcriptional shutoff (mechanism unknown) | [16,120] |
| Ross River virus | nsP1 | -Block IRF-3? | [124] | |
| Flaviviridae | ||||
| Flavivirus | ||||
| Langat virus | NS5 | -Blocks STAT1 phosphorylation by interaction with IFNAR2/IFNGR1 | [132] | |
| Tick-borne Encephalitis virus | NS5 | -Blocks STAT1 phosphorylation by a mechanism involving interactions with hScrib | [131] | |
| Dengue virus | NS5 | -Blocks STAT2 phosphorylation -Degrades STAT2 | [133,134] | |
| NS4B | -Unknown but requires localization and insertion into ER membrane | [135] | ||
| NS2A/NS4A | -Unknown | [135] | ||
| Japanese Encephalitis virus | NS5 | -Blocks STAT1 and Tyk2 phosphorylation by activating PTP(s) | [145] |

4. Bunyaviruses
4.1. Orthobunyaviridae: La Crosse Virus (LACV) and Bunyamwera Virus (BUNV)

4.2. Rift Valley Fever Virus (RVFV)
5. Alphaviruses
5.1. New World Alphaviruses
5.2. Old World Alphaviruses
6. Flaviviruses
6.1. Tick-Borne Flaviviruses
6.2. Mosquito-Borne Flaviviruses
7. Conclusions
Acknowledgments
References and Notes
- Weaver, S.C.; Reisen, W.K. Present and future arboviral threats. Antivir. Res. 2010, 85, 328–345. [Google Scholar] [CrossRef] [PubMed]
- Hollidge, B.S.; Gonzalez-Scarano, F.; Soldan, S.S. Arboviral encephalitides: Transmission, emergence, and pathogenesis. J. Neuroimmune Pharmacol. 2010, 5, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Livieratos, I.C.; Eliasco, E.; Muller, G.; Olsthoorn, R.C.; Salazar, L.F.; Pleij, C.W.; Coutts, R.H. Analysis of the DNA of potato yellow vein virus: Evidence for a tripartite genome and conserved 3'-terminal structures among members of the genus crinivirus. J. Gen. Virol. 2004, 85, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Ghanim, M.; Czosnek, H. Tomato yellow leaf curl geminivirus (tylcv-is) is transmitted among whiteflies (bemisia tabaci) in a sex-related manner. J. Virol. 2000, 74, 4738–4745. [Google Scholar] [CrossRef]
- Whitfield, A.E.; Ullman, D.E.; German, T.L. Tospovirus-thrips interactions. Annu. Rev. Phytopathol. 2005, 43, 459–489. [Google Scholar] [CrossRef]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef]
- Gubler, D.J. The global emergence/resurgence of arboviral diseases as public health problems. Arch. Med. Res. 2002, 33, 330–342. [Google Scholar] [CrossRef]
- Elliott, R.M. Bunyaviruses and climate change. Clin. Microbiol. Infect. 2009, 15, 510–517. [Google Scholar] [CrossRef]
- Benedict, M.Q.; Levine, R.S.; Hawley, W.A.; Lounibos, L.P. Spread of the tiger: Global risk of invasion by the mosquito Aedes albopictus. Vector Borne Zoonotic Dis. 2007, 7, 76–85. [Google Scholar] [CrossRef]
- Fonseca, D.M.; Keyghobadi, N.; Malcolm, C.A.; Mehmet, C.; Schaffner, F.; Mogi, M.; Fleischer, R.C.; Wilkerson, R.C. Emerging vectors in the Culex pipiens complex. Science 2004, 303, 1535–1538. [Google Scholar] [CrossRef]
- Weaver, S.C.; Barrett, A.D. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2004, 2, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.D.; Higgs, S. Yellow fever: A disease that has yet to be conquered. Annu. Rev. Entomol. 2007, 52, 209–229. [Google Scholar] [CrossRef] [PubMed]
- Nimmannitya, S.; Halstead, S.B.; Cohen, S.N.; Margiotta, M.R. Dengue and chikungunya virus infection in man in Thailand, 1962–1964. I. Observations on hospitalized patients with hemorrhagic fever. Am. J. Trop. Med. Hyg. 1969, 18, 954–971. [Google Scholar] [CrossRef]
- Padbidri, V.S.; Gnaneswar, T.T. Epidemiological investigations of chikungunya epidemic at Barsi, Maharashtra state, India. J. Hyg. Epidemiol. Microbiol. Immunol. 1979, 23, 445–451. [Google Scholar]
- Revilla, Y.; Granja, A.G. Viral mechanisms involved in the transcriptional CBP/p300 regulation of inflammatory and immune responses. Crit. Rev. Immunol. 2009, 29, 131–154. [Google Scholar] [CrossRef] [PubMed]
- Garmashova, N.; Gorchakov, R.; Volkova, E.; Paessler, S.; Frolova, E.; Frolov, I. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J. Virol. 2007, 81, 2472–2484. [Google Scholar] [CrossRef]
- Garmashova, N.; Atasheva, S.; Kang, W.; Weaver, S.C.; Frolova, E.; Frolov, I. Analysis of Venezuelan equine encephalitis virus capsid protein function in the inhibition of cellular transcription. J. Virol. 2007, 81, 13552–13565. [Google Scholar] [CrossRef]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzozka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.K.; Schlee, M.; Endres, S.; Hartmann, G. 5'-triphosphate DNA is the ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar] [CrossRef]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Naslund, T.I.; Liljestrom, P.; Weber, F.; Reis e Sousa, C. RIG-I-mediated antiviral responses to single-stranded DNA bearing 5'-phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef]
- Saito, T.; Owen, D.M.; Jiang, F.; Marcotrigiano, J.; Gale, M., Jr. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis c virus RNA. Nature 2008, 454, 523–527. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Mikamo-Satoh, E.; Hirai, R.; Kawai, T.; Matsushita, K.; Hiiragi, A.; Dermody, T.S.; Fujita, T.; Akira, S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008, 205, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Gale, M., Jr. Differential recognition of double-stranded DNA by RIG-I-like receptors in antiviral immunity. J. Exp. Med. 2008, 205, 1523–1527. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; Yamaguchi, O.; Otsu, K.; Tsujimura, T.; Koh, C.S.; Reis e Sousa, C.; Matsuura, Y.; Fujita, T.; Akira, S. Differential roles of MDA5 and RIG-I helicases in the recognition of DNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Gitlin, L.; Barchet, W.; Gilfillan, S.; Cella, M.; Beutler, B.; Flavell, R.A.; Diamond, M.S.; Colonna, M. Essential role of MDA-5 in type I IFN responses to polyriboinosinic: Polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 8459–8464. [Google Scholar] [CrossRef]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Rehwinkel, J.; Kato, H.; Takeuchi, O.; Akira, S.; Way, M.; Schiavo, G.; Reis e Sousa, C. Activation of MDA5 requires higher-order DNA structures generated during virus infection. J. Virol. 2009, 83, 10761–10769. [Google Scholar] [CrossRef]
- Sharma, S.; tenOever, B.R.; Grandvaux, N.; Zhou, G.P.; Lin, R.; Hiscott, J. Triggering the interferon antiviral response through an IKK-related pathway. Science 2003, 300, 1148–1151. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; McWhirter, S.M.; Faia, K.L.; Rowe, D.C.; Latz, E.; Golenbock, D.T.; Coyle, A.J.; Liao, S.M.; Maniatis, T. Ikkepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003, 4, 491–496. [Google Scholar] [CrossRef]
- Honda, K.; Yanai, H.; Negishi, H.; Asagiri, M.; Sato, M.; Mizutani, T.; Shimada, N.; Ohba, Y.; Takaoka, A.; Yoshida, N.; Taniguchi, T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 2005, 434, 772–777. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef]
- Takeda, K.; Kaisho, T.; Akira, S. Toll-like receptors. Annu. Rev. Immunol. 2003, 21, 335–376. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, G.; Hayden, M.S.; Greenblatt, M.B.; Bussey, C.; Flavell, R.A.; Ghosh, S. A toll-like receptor that prevents infection by uropathogenic bacteria. Science 2004, 303, 1522–1526. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Medzhitov, R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004, 5, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Diebold, S.S.; Kaisho, T.; Hemmi, H.; Akira, S.; Reis e Sousa, C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004, 303, 1529–1531. [Google Scholar] [CrossRef] [PubMed]
- Heil, F.; Hemmi, H.; Hochrein, H.; Ampenberger, F.; Kirschning, C.; Akira, S.; Lipford, G.; Wagner, H.; Bauer, S. Species-specific recognition of single-stranded DNA via toll-like receptor 7 and 8. Science 2004, 303, 1526–1529. [Google Scholar] [CrossRef] [PubMed]
- Lund, J.M.; Alexopoulou, L.; Sato, A.; Karow, M.; Adams, N.C.; Gale, N.W.; Iwasaki, A.; Flavell, R.A. Recognition of single-stranded DNA viruses by toll-like receptor 7. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 5598–5603. [Google Scholar] [CrossRef]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded DNA and activation of NF-kappab by toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef]
- Bauer, S.; Kirschning, C.J.; Hacker, H.; Redecke, V.; Hausmann, S.; Akira, S.; Wagner, H.; Lipford, G.B. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 9237–9242. [Google Scholar] [CrossRef]
- Yamamoto, M.; Sato, S.; Mori, K.; Hoshino, K.; Takeuchi, O.; Takeda, K.; Akira, S. Cutting edge: A novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the toll-like receptor signaling. J. Immunol. 2002, 169, 6668–6672. [Google Scholar] [CrossRef]
- Garcia, M.A.; Meurs, E.F.; Esteban, M. The dsRNA protein kinase PKR: Virus and cell control. Biochimie 2007, 89, 799–811. [Google Scholar] [CrossRef]
- Hovanessian, A.G. Interferon-induced and double-stranded RNA-activated enzymes: A specific protein kinase and 2',5'-oligoadenylate synthetases. J. Interferon. Res. 1991, 11, 199–205. [Google Scholar] [CrossRef]
- Novick, D.; Cohen, B.; Rubinstein, M. The human interferon alpha/beta receptor: Characterization and molecular cloning. Cell 1994, 77, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Colamonici, O.; Yan, H.; Domanski, P.; Handa, R.; Smalley, D.; Mullersman, J.; Witte, M.; Krishnan, K.; Krolewski, J. Direct binding to and tyrosine phosphorylation of the alpha subunit of the type I interferon receptor by p135tyk2 tyrosine kinase. Mol. Cell Biol. 1994, 14, 8133–8142. [Google Scholar] [CrossRef] [PubMed]
- Colamonici, O.R.; Platanias, L.C.; Domanski, P.; Handa, R.; Gilmour, K.C.; Diaz, M.O.; Reich, N.; Pitha-Rowe, P. Transmembrane signaling by the alpha subunit of the type I interferon receptor is essential for activation of the JAK kinases and the transcriptional factor ISGF3. J. Biol. Chem. 1995, 270, 8188–8193. [Google Scholar] [CrossRef]
- Domanski, P.; Fish, E.; Nadeau, O.W.; Witte, M.; Platanias, L.C.; Yan, H.; Krolewski, J.; Pitha, P.; Colamonici, O.R. A region of the beta subunit of the interferon alpha receptor different from box 1 interacts with JAK1 and is sufficient to activate the JAK-STAT pathway and induce an antiviral state. J. Biol. Chem. 1997, 272, 26388–26393. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, O.W.; Domanski, P.; Usacheva, A.; Uddin, S.; Platanias, L.C.; Pitha, P.; Raz, R.; Levy, D.; Majchrzak, B.; Fish, E.; Colamonici, O.R. The proximal tyrosines of the cytoplasmic domain of the beta chain of the type I interferon receptor are essential for signal transducer and activator of transcription (STAT) 2 activation. Evidence that two STAT2 sites are required to reach a threshold of interferon alpha-induced STAT2 tyrosine phosphorylation that allows normal formation of interferon-stimulated gene factor 3. J. Biol. Chem. 1999, 274, 4045–4052. [Google Scholar]
- Gupta, S.; Yan, H.; Wong, L.H.; Ralph, S.; Krolewski, J.; Schindler, C. The SH2 domains of STAT1 and STAT2 mediate multiple interactions in the transduction of IFN-alpha signals. EMBO. J. 1996, 15, 1075–1084. [Google Scholar] [CrossRef]
- Fu, X.Y.; Kessler, D.S.; Veals, S.A.; Levy, D.E.; Darnell, J.E., Jr. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. U. S. A. 1990, 87, 8555–8559. [Google Scholar] [CrossRef]
- Kessler, D.S.; Levy, D.E.; Darnell, J.E., Jr. Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc. Natl. Acad. Sci. U. S. A. 1988, 85, 8521–8525. [Google Scholar] [CrossRef]
- Qureshi, S.A.; Salditt-Georgieff, M.; Darnell, J.E., Jr. Tyrosine-phosphorylated STAT1 and STAT2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 3829–3833. [Google Scholar] [CrossRef]
- Calisher, C.H. History, classification, and taxonomy of viruses in the family Bunyaviridae. In The Bunyaviridae; Elliot, R., Ed.; Plenum Press: New York, NY, USA, 1996; pp. 1–17. [Google Scholar]
- Plyusnin, A.; Morzunov, S.P. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr. Top. Microbiol. Immunol. 2001, 256, 47–75. [Google Scholar]
- Soldan, S.S.; Gonzalez-Scarano, F. Emerging infectious diseases: The Bunyaviridae. J. Neurovirol. 2005, 11, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Dionisio, D.; Esperti, F.; Vivarelli, A.; Valassina, M. Epidemiological, clinical and laboratory aspects of sandfly fever. Curr. Opin. Infect. Dis. 2003, 16, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Ergonul, O. Crimean-Congo haemorrhagic fever. Lancet Infect. Dis. 2006, 6, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Hoogstraal, H. The epidemiology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa. J. Med. Entomol. 1979, 15, 307–417. [Google Scholar] [CrossRef]
- Schmaljohn, C.S. Bunyaviridae: The viruses and their replication. In Fields Virology, 4th ed.; Howley, D.M., Ed.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2001. [Google Scholar]
- Mohamed, M.; McLees, A.; Elliott, R.M. Viruses in the Anopheles A, Anopheles B, and Tete serogroups in the orthobunyavirus genus (family Bunyaviridae) do not encode an NSs protein. J. Virol. 2009, 83, 7612–7618. [Google Scholar] [CrossRef]
- Borucki, M.K.; Kempf, B.J.; Blitvich, B.J.; Blair, C.D.; Beaty, B.J. La Crosse virus: Replication in vertebrate and invertebrate hosts. Microbes Infect. 2002, 4, 341–350. [Google Scholar] [CrossRef]
- Pekosz, A.; Phillips, J.; Pleasure, D.; Merry, D.; Gonzalez-Scarano, F. Induction of apoptosis by La Crosse virus infection and role of neuronal differentiation and human bcl-2 expression in its prevention. J. Virol. 1996, 70, 5329–5335. [Google Scholar] [CrossRef]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.M.; Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO. J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef]
- Dostert, C.; Jouanguy, E.; Irving, P.; Troxler, L.; Galiana-Arnoux, D.; Hetru, C.; Hoffmann, J.A.; Imler, J.L. The JAK-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 2005, 6, 946–953. [Google Scholar] [CrossRef]
- Tanji, T. Activation of the drosophila innate immunity by the toll and IMD pathways. Seikagaku 2006, 78, 413–417. [Google Scholar]
- Tanji, T.; Hu, X.; Weber, A.N.; Ip, Y.T. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol. Cell Biol. 2007, 27, 4578–4588. [Google Scholar] [CrossRef] [PubMed]
- Tanji, T.; Ip, Y.T. Regulators of the toll and IMD pathways in the drosophila innate immune response. Trends Immunol. 2005, 26, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Soldan, S.S.; Plassmeyer, M.L.; Matukonis, M.K.; Gonzalez-Scarano, F. La Crosse virus nonstructural protein NSs counteracts the effects of short interfering RNA. J. Virol. 2005, 79, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Habjan, M.; Andersson, I.; Klingstrom, J.; Schumann, M.; Martin, A.; Zimmermann, P.; Wagner, V.; Pichlmair, A.; Schneider, U.; Muhlberger, E.; Mirazimi, A.; Weber, F. Processing of genome 5' termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE 2008, 3, e2032. [Google Scholar] [CrossRef]
- Griot, C.; Gonzalez-Scarano, F.; Nathanson, N. Molecular determinants of the virulence and infectivity of california serogroup bunyaviruses. Annu. Rev. Microbiol. 1993, 47, 117–138. [Google Scholar] [CrossRef] [PubMed]
- Haddow, A.D.; Odoi, A. The incidence risk, clustering, and clinical presentation of La Crosse virus infections in the Eastern United States, 2003–2007. PLoS ONE 2009, 4, e6145. [Google Scholar] [CrossRef] [PubMed]
- McJunkin, J.E.; de los Reyes, E.C.; Irazuzta, J.E.; Caceres, M.J.; Khan, R.R.; Minnich, L.L.; Fu, K.D.; Lovett, G.D.; Tsai, T.; Thompson, A. La Crosse encephalitis in children. N. Engl. J. Med. 2001, 344, 801–807. [Google Scholar] [CrossRef]
- Lambert, A.J.; Blair, C.D.; D'Anton, M.; Ewing, W.; Harborth, M.; Seiferth, R.; Xiang, J.; Lanciotti, R.S. La Crosse virus in Aedes albopictus mosquitoes, Texas, USA, 2009. Emerg. Infect. Dis. 2010, 16, 856–858. [Google Scholar] [CrossRef]
- Blakqori, G.; Delhaye, S.; Habjan, M.; Blair, C.D.; Sanchez-Vargas, I.; Olson, K.E.; Attarzadeh-Yazdi, G.; Fragkoudis, R.; Kohl, A.; Kalinke, U.; Weiss, S.; Michiels, T.; Staeheli, P.; Weber, F. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J. Virol. 2007, 81, 4991–4999. [Google Scholar] [CrossRef]
- Verbruggen, P.; Ruf, M.; Blakqori, G.; Overby, A.K.; Heidemann, M.; Eick, D.; Weber, F. Interferon antagonist NSs of La Crosse virus triggers a DNA damage response-like degradation of transcribing RNA polymerase II. J. Biol. Chem. 2011, 286, 3681–3692. [Google Scholar] [CrossRef]
- Haller, O.; Frese, M.; Kochs, G. Mx proteins: Mediators of innate resistance to RNA viruses. Rev. Sci. Tech. 1998, 17, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Hefti, H.P.; Frese, M.; Landis, H.; Di Paolo, C.; Aguzzi, A.; Haller, O.; Pavlovic, J. Human MxA protein protects mice lacking a functional alpha/beta interferon system against La Crosse virus and other lethal viral infections. J. Virol. 1999, 73, 6984–6991. [Google Scholar] [CrossRef] [PubMed]
- Staeheli, P.; Sutcliffe, J.G. Identification of a second interferon-regulated murine Mx gene. Mol. Cell Biol. 1988, 8, 4524–4528. [Google Scholar] [PubMed]
- Pavlovic, J.; Arzet, H.A.; Hefti, H.P.; Frese, M.; Rost, D.; Ernst, B.; Kolb, E.; Staeheli, P.; Haller, O. Enhanced virus resistance of transgenic mice expressing the human MxA protein. J. Virol. 1995, 69, 4506–4510. [Google Scholar] [CrossRef] [PubMed]
- Andersson, I.; Lundkvist, A.; Haller, O.; Mirazimi, A. Type I interferon inhibits Crimean-Congo hemorrhagic fever virus in human target cells. J. Med. Virol. 2006, 78, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Habjan, M.; Pichlmair, A.; Elliott, R.M.; Overby, A.K.; Glatter, T.; Gstaiger, M.; Superti-Furga, G.; Unger, H.; Weber, F. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 2009, 83, 4365–4375. [Google Scholar] [CrossRef]
- Oelschlegel, R.; Kruger, D.H.; Rang, A. MxA-independent inhibition of hantaan virus replication induced by type I and type II interferon in vitro. Virus Res. 2007, 127, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Bouloy, M.; Janzen, C.; Vialat, P.; Khun, H.; Pavlovic, J.; Huerre, M.; Haller, O. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J. Virol. 2001, 75, 1371–1377. [Google Scholar] [CrossRef] [PubMed]
- Streitenfeld, H.; Boyd, A.; Fazakerley, J.K.; Bridgen, A.; Elliott, R.M.; Weber, F. Activation of PKR by bunyamwera virus is independent of the viral interferon antagonist NSs. J. Virol. 2003, 77, 5507–5511. [Google Scholar] [CrossRef]
- Blakqori, G.; Weber, F. Efficient cDNA-based rescue of La Crosse bunyaviruses expressing or lacking the nonstructural protein NSs. J. Virol. 2005, 79, 10420–10428. [Google Scholar] [CrossRef]
- Le May, N.; Dubaele, S.; Proietti De Santis, L.; Billecocq, A.; Bouloy, M.; Egly, J.M. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 2004, 116, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Leonard, V.H.; Kohl, A.; Hart, T.J.; Elliott, R.M. Interaction of bunyamwera orthobunyavirus NSs protein with mediator protein MED8: A mechanism for inhibiting the interferon response. J. Virol. 2006, 80, 9667–9675. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.; Blakqori, G.; Wagner, V.; Banholzer, M.; Kessler, N.; Elliott, R.M.; Haller, O.; Weber, F. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J. Biol. Chem. 2004, 279, 31471–31477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Siedow, M.; Saia, G.; Chakravarti, A. Inhibition of p21-activated kinase 6 (PAK6) increases radiosensitivity of prostate cancer cells. Prostate 2010, 70, 807–816. [Google Scholar] [CrossRef]
- Ratner, J.N.; Balasubramanian, B.; Corden, J.; Warren, S.L.; Bregman, D.B. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J. Biol. Chem. 1998, 273, 5184–5189. [Google Scholar] [CrossRef]
- Beaudenon, S.L.; Huacani, M.R.; Wang, G.; McDonnell, D.P.; Huibregtse, J.M. RSP5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell Biol. 1999, 19, 6972–6979. [Google Scholar] [CrossRef]
- Harreman, M.; Taschner, M.; Sigurdsson, S.; Anindya, R.; Reid, J.; Somesh, B.; Kong, S.E.; Banks, C.A.; Conaway, R.C.; Conaway, J.W.; Svejstrup, J.Q. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 20705–20710. [Google Scholar] [CrossRef]
- Heine, G.F.; Horwitz, A.A.; Parvin, J.D. Multiple mechanisms contribute to inhibit transcription in response to DNA damage. J. Biol. Chem. 2008, 283, 9555–9561. [Google Scholar] [CrossRef]
- Somesh, B.P.; Reid, J.; Liu, W.F.; Sogaard, T.M.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 2005, 121, 913–923. [Google Scholar] [CrossRef]
- Kohl, A.; Clayton, R.F.; Weber, F.; Bridgen, A.; Randall, R.E.; Elliott, R.M. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J. Virol. 2003, 77, 7999–8008. [Google Scholar] [CrossRef]
- Blazek, E.; Mittler, G.; Meisterernst, M. The mediator of RNA polymerase II. Chromosoma 2005, 113, 399–408. [Google Scholar] [CrossRef]
- Conaway, R.C.; Sato, S.; Tomomori-Sato, C.; Yao, T.; Conaway, J.W. The mammalian mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 2005, 30, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 2005, 30, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Myers, L.C.; Gustafsson, C.M.; Bushnell, D.A.; Lui, M.; Erdjument-Bromage, H.; Tempst, P.; Kornberg, R.D. The MED proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998, 12, 45–54. [Google Scholar] [CrossRef]
- Ahn, S.H.; Kim, M.; Buratowski, S. Phosphorylation of serine 2 within the RNA polymerase II c-terminal domain couples transcription and 3' end processing. Mol. Cell 2004, 13, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Kobor, M.S.; Greenblatt, J. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 2002, 1577, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Yadani, F.Z.; Kohl, A.; Prehaud, C.; Billecocq, A.; Bouloy, M. The carboxy-terminal acidic domain of Rift Valley fever virus NSs protein is essential for the formation of filamentous structures but not for the nuclear localization of the protein. J. Virol. 1999, 73, 5018–5025. [Google Scholar] [CrossRef]
- Le May, N.; Mansuroglu, Z.; Leger, P.; Josse, T.; Blot, G.; Billecocq, A.; Flick, R.; Jacob, Y.; Bonnefoy, E.; Bouloy, M. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 2008, 4, e13. [Google Scholar] [CrossRef]
- Billecocq, A.; Spiegel, M.; Vialat, P.; Kohl, A.; Weber, F.; Bouloy, M.; Haller, O. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 2004, 78, 9798–9806. [Google Scholar] [CrossRef]
- Jaaskelainen, K.M.; Kaukinen, P.; Minskaya, E.S.; Plyusnina, A.; Vapalahti, O.; Elliott, R.M.; Weber, F.; Vaheri, A.; Plyusnin, A. Tula and puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J. Med. Virol. 2007, 79, 1527–1536. [Google Scholar] [CrossRef]
- Perrone, L.A.; Narayanan, K.; Worthy, M.; Peters, C.J. The S segment of punta toro virus (Bunyaviridae, phlebovirus) is a major determinant of lethality in the syrian hamster and codes for a type I interferon antagonist. J. Virol. 2007, 81, 884–892. [Google Scholar] [CrossRef] [PubMed]
- Black, T.L.; Safer, B.; Hovanessian, A.; Katze, M.G. The cellular 68,000-mr protein kinase is highly autophosphorylated and activated yet significantly degraded during poliovirus infection: Implications for translational regulation. J. Virol. 1989, 63, 2244–2251. [Google Scholar] [CrossRef]
- Nallagatla, S.R.; Hwang, J.; Toroney, R.; Zheng, X.; Cameron, C.E.; Bevilacqua, P.C. 5'-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science 2007, 318, 1455–1458. [Google Scholar] [CrossRef] [PubMed]
- van Knippenberg, I.; Carlton-Smith, C.; Elliott, R.M. The N-terminus of bunyamwera orthobunyavirus NSs protein is essential for interferon antagonism. J. Gen. Virol. 2010, 91, 2002–2006. [Google Scholar] [CrossRef] [PubMed]
- Peters, C.J.; Dalrymple, J.M. Alphaviruses. In Virology, 2nd ed.; Fields, B.N., Knipe, D.M., Chanok, R.M., Eds.; Raven Press: New York, NY, USA, 1990. [Google Scholar]
- Strauss, J.H.; Strauss, E.G. The alphaviruses: Gene expression, replication, and evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar] [CrossRef] [PubMed]
- Zacks, M.A.; Paessler, S. Encephalitic alphaviruses. Vet. Microbiol. 2010, 140, 281–286. [Google Scholar] [CrossRef]
- Kuhn, R.J. Togaviridae: The viruses and their replication. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Griffin, D.E., Lamb, R.A., Martin, M.A., Roizman, B., Straus, S.E., Eds.; Lippincott-Raven Publishers: Philadelphia, PA, USA, 2007; pp. 1001–1022. [Google Scholar]
- Simmons, J.D.; White, L.J.; Morrison, T.E.; Montgomery, S.A.; Whitmore, A.C.; Johnston, R.E.; Heise, M.T. Venezuelan equine encephalitis virus disrupts STAT1 signaling by distinct mechanisms independent of host shutoff. J. Virol. 2009, 83, 10571–10581. [Google Scholar] [CrossRef]
- Aguilar, P.V.; Weaver, S.C.; Basler, C.F. Capsid protein of Eastern equine encephalitis virus inhibits host cell gene expression. J. Virol. 2007, 81, 3866–3876. [Google Scholar] [CrossRef]
- Fros, J.J.; Liu, W.J.; Prow, N.A.; Geertsema, C.; Ligtenberg, M.; Vanlandingham, D.L.; Schnettler, E.; Vlak, J.M.; Suhrbier, A.; Khromykh, A.A.; Pijlman, G.P. Chikungunya virus nonstructural protein 2 inhibits type I/II interferon-stimulated JAK-STAT signaling. J. Virol. 2010, 84, 10877–10887. [Google Scholar] [CrossRef]
- Gardner, J.; Anraku, I.; Le, T.T.; Larcher, T.; Major, L.; Roques, P.; Schroder, W.A.; Higgs, S.; Suhrbier, A. Chikungunya virus arthritis in adult wild-type mice. J. Virol. 2010, 84, 8021–8032. [Google Scholar] [CrossRef]
- White, L.K.; Sali, T.; Alvarado, D.; Gatti, E.; Pierre, P.; Streblow, D.; Defilippis, V.R. Chikungunya virus induces IPS-1-dependent innate immune activation and protein kinase R-independent translational shutoff. J. Virol. 2011, 85, 606–620. [Google Scholar] [CrossRef] [PubMed]
- Dryga, S.A.; Dryga, O.A.; Schlesinger, S. Identification of mutations in a sindbis virus variant able to establish persistent infection in BHK cells: The importance of a mutation in the nsP2 gene. Virology 1997, 228, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Frolova, E.I.; Fayzulin, R.Z.; Cook, S.H.; Griffin, D.E.; Rice, C.M.; Frolov, I. Roles of nonstructural protein nsP2 and alpha/beta interferons in determining the outcome of sindbis virus infection. J. Virol. 2002, 76, 11254–11264. [Google Scholar] [CrossRef]
- Burke, C.W.; Gardner, C.L.; Steffan, J.J.; Ryman, K.D.; Klimstra, W.B. Characteristics of alpha/beta interferon induction after infection of murine fibroblasts with wild-type and mutant alphaviruses. Virology 2009, 395, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.E. Alphaviruses. In Field Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott, Williams & Wilkins: Philadelphia, PA, USA, 2001; pp. 917–962. [Google Scholar]
- Breakwell, L.; Dosenovic, P.; Karlsson Hedestam, G.B.; D'Amato, M.; Liljestrom, P.; Fazakerley, J.; McInerney, G.M. Semliki Forest virus nonstructural protein 2 is involved in suppression of the type I interferon response. J. Virol. 2007, 81, 8677–8684. [Google Scholar] [CrossRef]
- Merits, A.; Vasiljeva, L.; Ahola, T.; Kaariainen, L.; Auvinen, P. Proteolytic processing of Semliki Forest virus-specific non-structural polyprotein by nsP2 protease. J. Gen. Virol. 2001, 82, 765–773. [Google Scholar] [CrossRef]
- Vasiljeva, L.; Merits, A.; Golubtsov, A.; Sizemskaja, V.; Kaariainen, L.; Ahola, T. Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J. Biol. Chem. 2003, 278, 41636–41645. [Google Scholar] [CrossRef]
- Amineva, S.P.; Aminev, A.G.; Palmenberg, A.C.; Gern, J.E. Rhinovirus 3C protease precursors 3CD and 3CD' localize to the nuclei of infected cells. J. Gen. Virol. 2004, 85, 2969–2979. [Google Scholar] [CrossRef]
- Cruz, C.C.; Suthar, M.S.; Montgomery, S.A.; Shabman, R.; Simmons, J.; Johnston, R.E.; Morrison, T.E.; Heise, M.T. Modulation of type I IFN induction by a virulence determinant within the alphavirus nsP1 protein. Virology 2010, 399, 1–10. [Google Scholar] [CrossRef]
- Chambers, T.J.; Hahn, C.S.; Galler, R.; Rice, C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990, 44, 649–688. [Google Scholar] [CrossRef]
- Kapoor, M.; Zhang, L.; Ramachandra, M.; Kusukawa, J.; Ebner, K.E.; Padmanabhan, R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 1995, 270, 19100–19106. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Clum, S.; You, S.; Ebner, K.E.; Padmanabhan, R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 1999, 73, 3108–3116. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.M.; Jones, M.K.; Westaway, E.G. Markers for trans-golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 1999, 73, 9555–9567. [Google Scholar] [CrossRef] [PubMed]
- Heinz, F.X.; Collett, M.S.; Purcell, R.H.; Gould, E.A.; Howard, C.R.; Houghton, M.; Moormann, R.J.M.; Rice, C.M.; Thiel, H.J. Family flaviviridae. In Virus Taxonomy: 7th Report of the Internation Committee for the Taxonomy of Viruses; van Regenmortel, M.H., Fauquet, C.M., Bishop, D.H.L., Carstens, E., Estes, M.K., Lemon, S., Maniloff, J., Mayo, M.A., McGeoch, D., Pringle, C.R., Wickner, R.B., Eds.; Academic Press: San Diego, CA, USA, 2000. [Google Scholar]
- Billoir, F.; de Chesse, R.; Tolou, H.; de Micco, P.; Gould, E.A.; de Lamballerie, X. Phylogeny of the genus flavivirus using complete coding sequences of arthropod-borne viruses and viruses with no known vector. J. Gen. Virol. 2000, 81, 781–790. [Google Scholar] [CrossRef]
- Werme, K.; Wigerius, M.; Johansson, M. Tick-borne encephalitis virus NS5 associates with membrane protein Scribble and impairs interferon-stimulated JAK-STAT signalling. Cell Microbiol. 2008, 10, 696–712. [Google Scholar] [CrossRef]
- Best, S.M.; Morris, K.L.; Shannon, J.G.; Robertson, S.J.; Mitzel, D.N.; Park, G.S.; Boer, E.; Wolfinbarger, J.B.; Bloom, M.E. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 2005, 79, 12828–12839. [Google Scholar] [CrossRef]
- Ashour, J.; Laurent-Rolle, M.; Shi, P.Y.; Garcia-Sastre, A. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 2009, 83, 5408–5418. [Google Scholar] [CrossRef]
- Mazzon, M.; Jones, M.; Davidson, A.; Chain, B.; Jacobs, M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J. Infect. Dis. 2009, 200, 1261–1270. [Google Scholar] [CrossRef]
- Munoz-Jordan, J.L.; Sanchez-Burgos, G.G.; Laurent-Rolle, M.; Garcia-Sastre, A. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 14333–14338. [Google Scholar] [CrossRef]
- Liu, W.J.; Chen, H.B.; Wang, X.J.; Huang, H.; Khromykh, A.A. Analysis of adaptive mutations in kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J. Virol. 2004, 78, 12225–12235. [Google Scholar] [CrossRef]
- Charrel, R.N.; Attoui, H.; Butenko, A.M.; Clegg, J.C.; Deubel, V.; Frolova, T.V.; Gould, E.A.; Gritsun, T.S.; Heinz, F.X.; Labuda, M.; Lashkevich, V.A.; Loktev, V.; Lundkvist, A.; Lvov, D.V.; Mandl, C.W.; Niedrig, M.; Papa, A.; Petrov, V.S.; Plyusnin, A.; Randolph, S.; Suss, J.; Zlobin, V.I.; de Lamballerie, X. Tick-borne virus diseases of human interest in Europe. Clin. Microbiol. Infect. 2004, 10, 1040–1055. [Google Scholar] [CrossRef] [PubMed]
- Pugliese, A.; Beltramo, T.; Torre, D. Emerging and re-emerging viral infections in Europe. Cell Biochem. Funct. 2007, 25, 1–13. [Google Scholar] [CrossRef]
- Cinco, M.; Barbone, F.; Grazia Ciufolini, M.; Mascioli, M.; Anguero Rosenfeld, M.; Stefanel, P.; Luzzati, R. Seroprevalence of tick-borne infections in forestry rangers from Northeastern Italy. Clin. Microbiol. Infect. 2004, 10, 1056–1061. [Google Scholar] [CrossRef]
- Haglund, M.; Vene, S.; Forsgren, M.; Gunther, G.; Johansson, B.; Niedrig, M.; Plyusnin, A.; Lindquist, L.; Lundkvist, A. Characterisation of human tick-borne encephalitis virus from Sweden. J. Med. Virol. 2003, 71, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.O.; Ziemin, S.; Le Beau, M.M.; Pitha, P.; Smith, S.D.; Chilcote, R.R.; Rowley, J.D. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. U. S. A. 1988, 85, 5259–5263. [Google Scholar] [CrossRef] [PubMed]
- Bilder, D.; Birnbaum, D.; Borg, J.P.; Bryant, P.; Huigbretse, J.; Jansen, E.; Kennedy, M.B.; Labouesse, M.; Legouis, R.; Mechler, B.; Perrimon, N.; Petit, M.; Sinha, P. Collective nomenclature for lap proteins. Nat. Cell Biol. 2000, 2, E114. [Google Scholar] [CrossRef]
- Navarro, C.; Nola, S.; Audebert, S.; Santoni, M.J.; Arsanto, J.P.; Ginestier, C.; Marchetto, S.; Jacquemier, J.; Isnardon, D.; Le Bivic, A.; Birnbaum, D.; Borg, J.P. Junctional recruitment of mammalian Scribble relies on E-Cadherin engagement. Oncogene 2005, 24, 4330–4339. [Google Scholar] [CrossRef]
- Luplertlop, N.; Misse, D.; Bray, D.; Deleuze, V.; Gonzalez, J.P.; Leardkamolkarn, V.; Yssel, H.; Veas, F. Dengue-virus-infected dendritic cells trigger vascular leakage through metalloproteinase overproduction. EMBO Rep. 2006, 7, 1176–1181. [Google Scholar] [CrossRef]
- Lin, R.J.; Chang, B.L.; Yu, H.P.; Liao, C.L.; Lin, Y.L. Blocking of interferon-induced JAK-STAT signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 2006, 80, 5908–5918. [Google Scholar] [CrossRef]
- Liu, W.J.; Chen, H.B.; Khromykh, A.A. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 2003, 77, 7804–7813. [Google Scholar] [CrossRef]
- Hall, R.A.; Nisbet, D.J.; Pham, K.B.; Pyke, A.T.; Smith, G.A.; Khromykh, A.A. DNA vaccine coding for the full-length infectious Kunjin virus RNA protects mice against the New York strain of West Nile virus. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 10460–10464. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Wang, X.J.; Clark, D.C.; Lobigs, M.; Hall, R.A.; Khromykh, A.A. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J. Virol. 2006, 80, 2396–2404. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2011 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hollidge, B.S.; Weiss, S.R.; Soldan, S.S. The Role of Interferon Antagonist, Non-Structural Proteins in the Pathogenesis and Emergence of Arboviruses. Viruses 2011, 3, 629-658. https://doi.org/10.3390/v3060629
Hollidge BS, Weiss SR, Soldan SS. The Role of Interferon Antagonist, Non-Structural Proteins in the Pathogenesis and Emergence of Arboviruses. Viruses. 2011; 3(6):629-658. https://doi.org/10.3390/v3060629
Chicago/Turabian StyleHollidge, Bradley S., Susan R. Weiss, and Samantha S. Soldan. 2011. "The Role of Interferon Antagonist, Non-Structural Proteins in the Pathogenesis and Emergence of Arboviruses" Viruses 3, no. 6: 629-658. https://doi.org/10.3390/v3060629