Influenza B-Cells Protective Epitope Characterization: A Passkey for the Rational Design of New Broad-Range Anti-Influenza Vaccines
Abstract
:1. Introduction
2. Hemagglutinin and Protective mAbs
3. Broadly Neutralizing mAbs: Dual Role in the Fight Against a Variable Virus
 |
3.1. “Classical” Vaccine Limitations
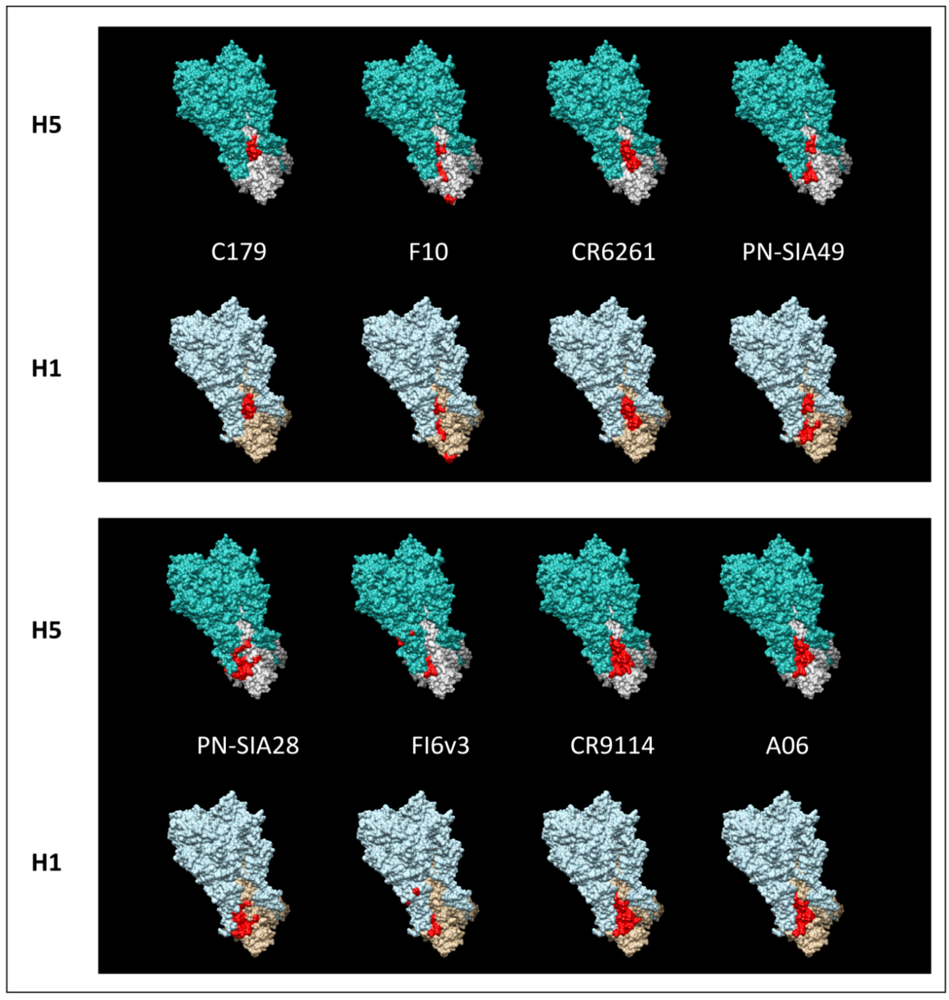
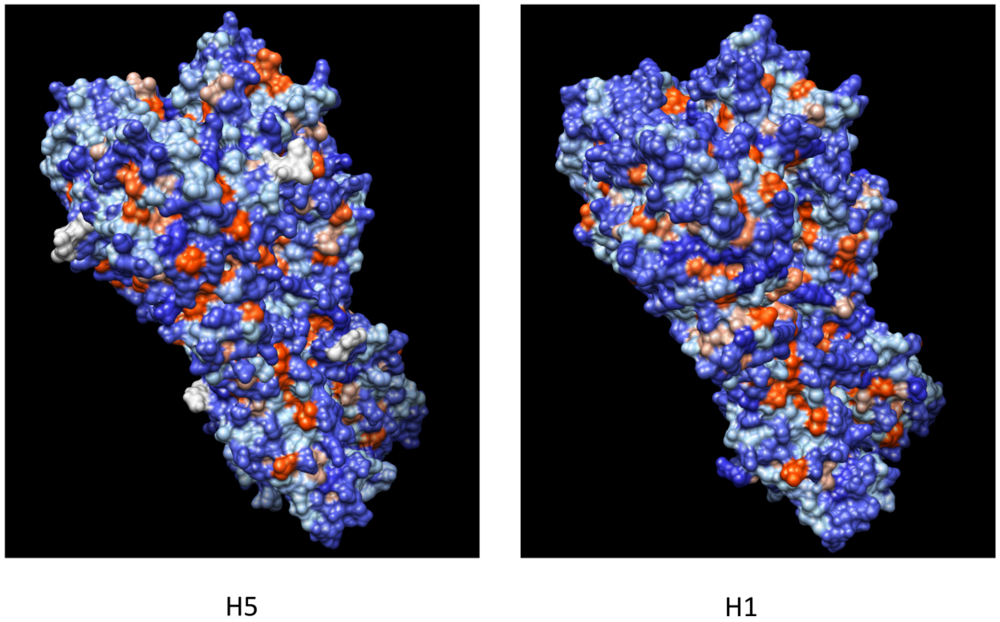
5. H5N1 Ccross-Clade Protection
6. Conclusions
Conflict of Interest
References
- Kawaoka, Y. H5N1: Flu transmission work is urgent. Nature 2012, 482, 155. [Google Scholar]
- Osterholm, M.T.; Henderson, D.A. Public health and biosecurity. Life sciences at a crossroads: Respiratory transmissible H5N1. Science 2012, 335, 801–802. [Google Scholar] [CrossRef]
- Fouchier, R.; Osterhaus, A.B.; Steinbruner, J.; Yuen, K.Y.; Henderson, D.A.; Klotz, L.; Sylvester, E.; Taubenberger, J.K.; Ebright, R.H.; Heymann, D.L. Preventing pandemics: The fight over flu. Nature 2012, 481, 257–259. [Google Scholar]
- Subbarao, K.; Katz, J. Avian influenza viruses infecting humans. Cellular and molecular life sciences : CMLS 2000, 57, 1770–1784. [Google Scholar] [CrossRef]
- Steel, J. New strategies for the development of H5N1 subtype influenza vaccines: progress and challenges. BioDrugs 2011, 25, 285–298. [Google Scholar]
- Imai, M.; Watanabe, T.; Hatta, M.; Das, S.C.; Ozawa, M.; Shinya, K.; Zhong, G.; Hanson, A.; Katsura, H.; Watanabe, S.; Li, C.; Kawakami, E.; Yamada, S.; Kiso, M.; Suzuki, Y.; Maher, E.A.; Neumann, G.; Kawaoka, Y. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 2012, 486, 420–428. [Google Scholar]
- Osterholm, M.T.; Kelley, N.S. Mammalian-transmissible H5N1 influenza: facts and perspective. mBio 2012, 3, e00045–00012. [Google Scholar]
- Yong, E. Influenza: Five questions on H5N1. Nature 2012, 486, 456–458. [Google Scholar] [CrossRef]
- Vergara-Jaque, A.; Poblete, H.; Lee, E.H.; Schulten, K.; Gonzalez-Nilo, F.D.; Chipot, C.J. Molecular basis of drug resistance in A/H1N1 virus. J. Chem. Inf. Model 2012. [Google Scholar]
- Hurt, A.C.; Chotpitayasunondh, T.; Cox, N.J.; Daniels, R.; Fry, A.M.; Gubareva, L.V.; Hayden, F.G.; Hui, D.S.; Hungnes, O.; Lackenby, A.; Lim, W.; Meijer, A.; Penn, C.; Tashiro, M.; Uyeki, T.M.; Zambon, M. Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect. Dis. 2012, 12, 240–248. [Google Scholar] [CrossRef]
- Redlberger-Fritz, M.; Aberle, S.W.; Strassl, R.; Popow-Kraupp, T. Rapid identification of neuraminidase inhibitor resistance mutations in seasonal influenza virus A(H1N1), A(H1N1)2009, and A(H3N2) subtypes by melting point analysis. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1593–1601. [Google Scholar]
- Ilyushina, N.A.; Bovin, N.V.; Webster, R.G. Decreased neuraminidase activity is important for the adaptation of H5N1 influenza virus to human airway epithelium. J. Virol. 2012, 86, 4724–4733. [Google Scholar]
- Blondel, B.; Mahjoub, N.; Drewniak, N.; Launay, O.; Goffinet, F. Failure of the vaccination campaign against A(H1N1) influenza in pregnant women in France: Results from a national survey. Vaccine 2012, 30, 5661–5665. [Google Scholar] [CrossRef] [Green Version]
- Nachtnebel, M.; Greutelaers, B.; Falkenhorst, G.; Jorgensen, P.; Dehnert, M.; Schweiger, B.; Trader, C.; Buda, S.; Eckmanns, T.; Wichmann, O.; Hellenbrand, W. Lessons from a one-year hospital-based surveillance of acute respiratory infections in Berlin- comparing case definitions to monitor influenza. BMC Public Health 2012, 12, 245. [Google Scholar] [CrossRef]
- Olivier, C.W. Influenza vaccination coverage rate in children: reasons for a failure and how to go forward. Hum. Vaccin. Immunother. 2012, 8, 107–118. [Google Scholar] [CrossRef]
- Burioni, R.; Canducci, F.; Clementi, M. Pregnancy and H1N1 infection. Lancet 2009, 374, 1417, author reply 1417-1418. [Google Scholar]
- Zhou, F.; Zhou, J.; Ma, L.; Song, S.; Zhang, X.; Li, W.; Jiang, S.; Wang, Y.; Liao, G. High-yield production of a stable Vero cell-based vaccine candidate against the highly pathogenic avian influenza virus H5N1. Biochem. Biophys. Res. Commun. 2012, 421, 850–854. [Google Scholar] [CrossRef]
- Zhou, F.; Wang, G.; Buchy, P.; Cai, Z.; Chen, H.; Chen, Z.; Cheng, G.; Wan, X.F.; Deubel, V.; Zhou, P. A triclade DNA vaccine designed on the basis of a comprehensive serologic study elicits neutralizing antibody responses against all clades and subclades of highly pathogenic avian influenza H5N1 viruses. J. Virol. 2012, 86, 6970–6978. [Google Scholar] [CrossRef]
- Yang, P.; Duan, Y.; Zhang, P.; Li, Z.; Wang, C.; Dong, M.; Tang, C.; Xing, L.; Gu, H.; Zhao, Z.; Liu, X.; Zhang, S.; Wang, X. Multiple-clade H5N1 influenza split vaccine elicits broad cross protection against lethal influenza virus challenge in mice by intranasal vaccination. PloS One 2012, 7, e30252. [Google Scholar]
- Wodal, W.; Falkner, F.G.; Kerschbaum, A.; Gaiswinkler, C.; Fritz, R.; Kiermayr, S.; Portsmouth, D.; Savidis-Dacho, H.; Coulibaly, S.; Piskernik, C.; Hohenadl, C.; Howard, M.K.; Kistner, O.; Barrett, P.N.; Kreil, T.R. A cell culture-derived whole-virus H9N2 vaccine induces high titer antibodies against hemagglutinin and neuraminidase and protects mice from severe lung pathology and weight loss after challenge with a highly virulent H9N2 isolate. Vaccine 2012, 30, 4625–4631. [Google Scholar]
- Wang, B.Z.; Gill, H.S.; Kang, S.M.; Wang, L.; Wang, Y.C.; Vassilieva, E.V.; Compans, R.W. Enhanced influenza virus-like particle vaccines containing the extracellular domain of matrix protein 2 and a toll-like receptor ligand. Clin. Vaccine. Immunol. 2012, 19, 1119–1125. [Google Scholar] [CrossRef]
- Wang, B.; Yu, H.; Yang, F.R.; Huang, M.; Ma, J.H.; Tong, G.Z. Protective efficacy of a broadly cross-reactive swine influenza DNA vaccine encoding M2e, cytotoxic T lymphocyte epitope and consensus H3 hemagglutinin. Virol. J. 2012, 9, 127. [Google Scholar] [CrossRef]
- Underwood, P.A. Mapping of antigenic changes in the haemagglutinin of Hong Kong influenza (H3N2) strains using a large panel of monoclonal antibodies. J. Gen. Virol. 1982, 62 Pt 1, 153–169. [Google Scholar] [CrossRef]
- Wiley, D.C.; Wilson, I.A.; Skehel, J.J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 1981, 289, 373–378. [Google Scholar] [CrossRef]
- Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 1981, 289, 366–373. [Google Scholar] [CrossRef]
- Caton, A.J.; Brownlee, G.G.; Yewdell, J.W.; Gerhard, W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 1982, 31, 417–427. [Google Scholar] [CrossRef]
- Tsuchiya, E.; Sugawara, K.; Hongo, S.; Matsuzaki, Y.; Muraki, Y.; Li, Z.N.; Nakamura, K. Antigenic structure of the haemagglutinin of human influenza A/H2N2 virus. J. Gen. Virol. 2001, 82, 2475–2484. [Google Scholar]
- Kaverin, N.V.; Rudneva, I.A.; Ilyushina, N.A.; Varich, N.L.; Lipatov, A.S.; Smirnov, Y.A.; Govorkova, E.A.; Gitelman, A.K.; Lvov, D.K.; Webster, R.G. Structure of antigenic sites on the haemagglutinin molecule of H5 avian influenza virus and phenotypic variation of escape mutants. J. Gen. Virol. 2002, 83, 2497–2505. [Google Scholar]
- Kaverin, N.V.; Rudneva, I.A.; Govorkova, E.A.; Timofeeva, T.A.; Shilov, A.A.; Kochergin-Nikitsky, K.S.; Krylov, P.S.; Webster, R.G. Epitope mapping of the hemagglutinin molecule of a highly pathogenic H5N1 influenza virus by using monoclonal antibodies. J. Virol. 2007, 81, 12911–12917. [Google Scholar]
- Kaverin, N.V.; Rudneva, I.A.; Ilyushina, N.A.; Lipatov, A.S.; Krauss, S.; Webster, R.G. Structural differences among hemagglutinins of influenza A virus subtypes are reflected in their antigenic architecture: analysis of H9 escape mutants. J. Virol. 2004, 78, 240–249. [Google Scholar]
- Mancini, N.; Solforosi, L.; Clementi, N.; De Marco, D.; Clementi, M.; Burioni, R. A potential role for monoclonal antibodies in prophylactic and therapeutic treatment of influenza. Antiviral Res. 2011, 92, 15–26. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Kashyap, A.K.; Steel, J.; Rubrum, A.; Bhabha, G.; Khayat, R.; Lee, J.H.; Dillon, M.A.; O'Neil, R.E.; Faynboym, A.M.; Horowitz, M.; Horowitz, L.; Ward, A.B.; Palese, P.; Webby, R.; Lerner, R.A.; Bhatt, R.R.; Wilson, I.A. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 2012. [Google Scholar]
- Ohshima, N.; Iba, Y.; Kubota-Koketsu, R.; Asano, Y.; Okuno, Y.; Kurosawa, Y. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J. Virol. 2011, 85, 11048–11057. [Google Scholar] [CrossRef]
- Russell, R.J.; Kerry, P.S.; Stevens, D.J.; Steinhauer, D.A.; Martin, S.R.; Gamblin, S.J.; Skehel, J.J. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc. Natl. Acad. Sci. USA 2008, 105, 17736–17741. [Google Scholar]
- Haynes, B.F.; Fleming, J.; St Clair, E.W.; Katinger, H.; Stiegler, G.; Kunert, R.; Robinson, J.; Scearce, R.M.; Plonk, K.; Staats, H.F.; Ortel, T.L.; Liao, H.X.; Alam, S.M. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 2005, 308, 1906–1908. [Google Scholar]
- Cassady-Cain, R.L.; Kaushik, A.K. Increased negative selection impairs neonatal B cell repertoire but does not directly lead to generation of disease-associated IgM auto-antibodies. Int. Immunol. 2006, 18, 661–669. [Google Scholar] [CrossRef]
- Montefiori, D.; Sattentau, Q.; Flores, J.; Esparza, J.; Mascola, J. Antibody-based HIV-1 vaccines: recent developments and future directions. PLoS Medicine 2007, 4, e348. [Google Scholar] [CrossRef]
- Zhang, M.; Zharikova, D.; Mozdzanowska, K.; Otvos, L.; Gerhard, W. Fine specificity and sequence of antibodies directed against the ectodomain of matrix protein 2 of influenza A virus. Mol. Immunol. 2006, 43, 2195–2206. [Google Scholar] [CrossRef]
- Zharikova, D.; Mozdzanowska, K.; Feng, J.; Zhang, M.; Gerhard, W. Influenza type A virus escape mutants emerge in vivo in the presence of antibodies to the ectodomain of matrix protein 2. J. Virol. 2005, 79, 6644–6654. [Google Scholar]
- Wei, G.; Meng, W.; Guo, H.; Pan, W.; Liu, J.; Peng, T.; Chen, L.; Chen, C.Y. Potent neutralization of influenza A virus by a single-domain antibody blocking M2 ion channel protein. PloS One 2011, 6, e28309. [Google Scholar]
- Wang, Y.; Xu, H.; Wu, N.; Shi, H.; Wang, X.; Wang, T. Monoclonal antibody, but not synthetic peptide, targeting the ectodomain of influenza B virus M2 proton channel has antiviral activity. New Microbiol. 2010, 33, 311–317. [Google Scholar]
- Wang, Y.; Zhou, L.; Shi, H.; Xu, H.; Yao, H.; Xi, X.G.; Toyoda, T.; Wang, X.; Wang, T. Monoclonal antibody recognizing SLLTEVET epitope of M2 protein potently inhibited the replication of influenza A viruses in MDCK cells. Biochem. Biophys. Res. Commun. 2009, 385, 118–122. [Google Scholar] [CrossRef]
- Gabbard, J.; Velappan, N.; Di Niro, R.; Schmidt, J.; Jones, C.A.; Tompkins, S.M.; Bradbury, A.R. A humanized anti-M2 scFv shows protective in vitro activity against influenza. Protein Eng. Des. Sel. 2009, 22, 189–198. [Google Scholar]
- Fu, T.M.; Freed, D.C.; Horton, M.S.; Fan, J.; Citron, M.P.; Joyce, J.G.; Garsky, V.M.; Casimiro, D.R.; Zhao, Q.; Shiver, J.W.; Liang, X. Characterizations of four monoclonal antibodies against M2 protein ectodomain of influenza A virus. Virology 2009, 385, 218–226. [Google Scholar] [CrossRef]
- Beerli, R.R.; Bauer, M.; Schmitz, N.; Buser, R.B.; Gwerder, M.; Muntwiler, S.; Renner, W.A.; Saudan, P.; Bachmann, M.F. Prophylactic and therapeutic activity of fully human monoclonal antibodies directed against influenza A M2 protein. Virol. J. 2009, 6, 224. [Google Scholar]
- Zou, P.; Liu, W.; Wu, F.; Chen, Y.H. Fine-epitope mapping of an antibody that binds the ectodomain of influenza matrix protein 2. FEMS Immunol. Med. Microbiol. 2008, 53, 79–84. [Google Scholar] [CrossRef]
- Liu, W.; Zou, P.; Chen, Y.H. Monoclonal antibodies recognizing EVETPIRN epitope of influenza A virus M2 protein could protect mice from lethal influenza A virus challenge. Immunol. Lett. 2004, 93, 131–136. [Google Scholar] [CrossRef]
- Shoji, Y.; Chichester, J.A.; Palmer, G.A.; Farrance, C.E.; Stevens, R.; Stewart, M.; Goldschmidt, L.; Deyde, V.; Gubareva, L.; Klimov, A.; Mett, V.; Yusibov, V. An influenza N1 neuraminidase-specific monoclonal antibody with broad neuraminidase inhibition activity against H5N1 HPAI viruses. Hum. Vacc. 2011, 7 Suppl, 199–204. [Google Scholar]
- Ilyushina, N.; Rudneva, I.; Gambaryan, A.; Bovin, N.; Kaverin, N. Monoclonal antibodies differentially affect the interaction between the hemagglutinin of H9 influenza virus escape mutants and sialic receptors. Virology 2004, 329, 33–39. [Google Scholar] [CrossRef]
- Lee, J.T.; Air, G.M. Contacts between influenza virus N9 neuraminidase and monoclonal antibody NC10. Virology 2002, 300, 255–268. [Google Scholar] [CrossRef]
- Burioni, R.; Canducci, F.; Mancini, N.; Clementi, N.; Sassi, M.; De Marco, D.; Diotti, R.A.; Saita, D.; Sampaolo, M.; Sautto, G.; Pianezze, M.; Clementi, M. Monoclonal antibodies isolated from human B cells neutralize a broad range of H1 subtype influenza A viruses including swine-origin Influenza virus (S-OIV). Virology 2010, 399, 144–152. [Google Scholar] [CrossRef]
- Burioni, R.; Canducci, F.; Mancini, N.; Clementi, N.; Sassi, M.; De Marco, D.; Saita, D.; Diotti, R.A.; Sautto, G.; Sampaolo, M.; Clementi, M. Molecular cloning of the first human monoclonal antibodies neutralizing with high potency swine-origin influenza A pandemic virus (S-OIV). New Microbiol. 2009, 32, 319–324. [Google Scholar]
- Clementi, N.; De Marco, D.; Mancini, N.; Solforosi, L.; Moreno, G.J.; Gubareva, L.V.; Mishin, V.; Di Pietro, A.; Vicenzi, E.; Siccardi, A.G.; Clementi, M.; Burioni, R. A human monoclonal antibody with neutralizing activity against highly divergent influenza subtypes. PloS One 2011, 6, e28001. [Google Scholar]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; Silacci, C.; Fernandez-Rodriguez, B.M.; Agatic, G.; Bianchi, S.; Giacchetto-Sasselli, I.; Calder, L.; Sallusto, F.; Collins, P.; Haire, L.F.; Temperton, N.; Langedijk, J.P.; Skehel, J.J.; Lanzavecchia, A. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011, 333, 850–856. [Google Scholar]
- De Marco, D.; Clementi, N.; Mancini, N.; Solforosi, L.; Moreno, G.J.; Sun, X.; Tumpey, T.M.; Gubareva, L.V.; Mishin, V.; Clementi, M.; Burioni, R. A non-VH1-69 heterosubtypic neutralizing human monoclonal antibody protects mice against H1N1 and H5N1 viruses. PloS One 2012, 7, e34415. [Google Scholar]
- Dreyfus, C.; Laursen, N.S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongeneelen, M.; van der Vlugt, R.; Lamrani, M.; Korse, H.J.; Geelen, E.; Sahin, O.; Sieuwerts, M.; Brakenhoff, J.P.; Vogels, R.; Li, O.T.; Poon, L.L.; Peiris, M.; Koudstaal, W.; Ward, A.B.; Wilson, I.A.; Goudsmit, J.; Friesen, R.H. Highly conserved protective epitopes on influenza B viruses. Science 2012, 337, 1343–1348. [Google Scholar]
- Kashyap, A.K.; Steel, J.; Oner, A.F.; Dillon, M.A.; Swale, R.E.; Wall, K.M.; Perry, K.J.; Faynboym, A.; Ilhan, M.; Horowitz, M.; Horowitz, L.; Palese, P.; Bhatt, R.R.; Lerner, R.A. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc. Natl. Acad. Sci. USA 2008, 105, 5986–5991. [Google Scholar]
- Kashyap, A.K.; Steel, J.; Rubrum, A.; Estelles, A.; Briante, R.; Ilyushina, N.A.; Xu, L.; Swale, R.E.; Faynboym, A.M.; Foreman, P.K.; Horowitz, M.; Horowitz, L.; Webby, R.; Palese, P.; Lerner, R.A.; Bhatt, R.R. Protection from the 2009 H1N1 pandemic influenza by an antibody from combinatorial survivor-based libraries. PLoS Pathogens 2010, 6, e1000990. [Google Scholar]
- Okuno, Y.; Isegawa, Y.; Sasao, F.; Ueda, S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 1993, 67, 2552–2558. [Google Scholar]
- Throsby, M.; van den Brink, E.; Jongeneelen, M.; Poon, L.L.; Alard, P.; Cornelissen, L.; Bakker, A.; Cox, F.; van Deventer, E.; Guan, Y.; Cinatl, J.; ter Meulen, J.; Lasters, I.; Carsetti, R.; Peiris, M.; de Kruif, J.; Goudsmit, J. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PloS One 2008, 3, e3942. [Google Scholar]
- Sui, J.; Hwang, W.C.; Perez, S.; Wei, G.; Aird, D.; Chen, L.M.; Santelli, E.; Stec, B.; Cadwell, G.; Ali, M.; Wan, H.; Murakami, A.; Yammanuru, A.; Han, T.; Cox, N.J.; Bankston, L.A.; Donis, R.O.; Liddington, R.C.; Marasco, W.A. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009, 16, 265–273. [Google Scholar]
- Pos, W.; Luken, B.M.; Hovinga, J.A.; Turenhout, E.A.; Scheiflinger, F.; Dong, J.F.; Fijnheer, R.; Voorberg, J. VH1-69 germline encoded antibodies directed towards ADAMTS13 in patients with acquired thrombotic thrombocytopenic purpura. J. Thromb. Haemost. 2009, 7, 421–428. [Google Scholar] [CrossRef]
- Perotti, M.; Ghidoli, N.; Altara, R.; Diotti, R.A.; Clementi, N.; De Marco, D.; Sassi, M.; Clementi, M.; Burioni, R.; Mancini, N. Hepatitis C virus (HCV)-driven stimulation of subfamily-restricted natural IgM antibodies in mixed cryoglobulinemia. Autoimm. Rev. 2008, 7, 468–472. [Google Scholar] [CrossRef]
- Carbonari, M.; Caprini, E.; Tedesco, T.; Mazzetta, F.; Tocco, V.; Casato, M.; Russo, G.; Fiorilli, M. Hepatitis C virus drives the unconstrained monoclonal expansion of VH1-69-expressing memory B cells in type II cryoglobulinemia: a model of infection-driven lymphomagenesis. J. Immunol. 2005, 174, 6532–6539. [Google Scholar]
- Sautto, G.; Mancini, N.; Solforosi, L.; Diotti, R.A.; Clementi, M.; Burioni, R. HCV proteins and immunoglobulin variable gene (IgV) subfamilies in HCV-induced type II mixed cryoglobulinemia: a concurrent pathogenetic role. Clin. Dev. Immunol. 2012, 2012, 705013. [Google Scholar]
- Haynes, L.M.; Caidi, H.; Radu, G.U.; Miao, C.; Harcourt, J.L.; Tripp, R.A.; Anderson, L.J. Therapeutic monoclonal antibody treatment targeting respiratory syncytial virus (RSV) G protein mediates viral clearance and reduces the pathogenesis of RSV infection in BALB/c mice. J. Infect. Dis. 2009, 200, 439–447. [Google Scholar] [CrossRef]
- Burioni, R.; Williamson, R.A.; Sanna, P.P.; Bloom, F.E.; Burton, D.R. Recombinant human Fab to glycoprotein D neutralizes infectivity and prevents cell-to-cell transmission of herpes simplex viruses 1 and 2 in vitro. Proc. Natl. Acad. Sci. USA 1994, 91, 355–359. [Google Scholar] [CrossRef]
- Burioni, R.; Plaisant, P.; Manzin, A.; Rosa, D.; Delli Carri, V.; Bugli, F.; Solforosi, L.; Abrignani, S.; Varaldo, P.E.; Fadda, G.; Clementi, M. Dissection of human humoral immune response against hepatitis C virus E2 glycoprotein by repertoire cloning and generation of recombinant Fab fragments. Hepatology 1998, 28, 810–814. [Google Scholar] [CrossRef]
- Cabral, T.M.; Berhane, Y.; Schmidt, L.; Tracz, D.M.; Hole, K.; Leith, M.; Corbett, C.R. Development and characterization of neutralizing monoclonal antibodies against the pandemic H1N1 virus (2009). J. Virol. Methods 2012, 183, 25–33. [Google Scholar] [CrossRef]
- Burioni, R.; Perotti, M.; Mancini, N.; Clementi, M. Perspectives for the utilization of neutralizing human monoclonal antibodies as anti-HCV drugs. J. Hepatol. 2008, 49, 299–300. [Google Scholar]
- Clementi, N.; Mancini, N.; Solforosi, L.; Castelli, M.; Clementi, M.; Burioni, R. Phage Display-based Strategies for Cloning and Optimization of Monoclonal Antibodies Directed against Human Pathogens. IJMS 2012, 13, 8273–8292. [Google Scholar] [CrossRef]
- Plaisant, P.; Burioni, R.; Manzin, A.; Solforosi, L.; Candela, M.; Gabrielli, A.; Fadda, G.; Clementi, M. Human monoclonal recombinant Fabs specific for HCV antigens obtained by repertoire cloning in phage display combinatorial vectors. Res. Virol. 1997, 148, 165–169. [Google Scholar] [CrossRef]
- Burioni, R.; Plaisant, P.; Delli Carri, V.; Vannini, A.; Spanu, T.; Clementi, M.; Fadda, G.; Varaldo, P.E. An improved phage display vector for antibody repertoire cloning by construction of combinatorial libraries. Res. Virol. 1997, 148, 161–164. [Google Scholar] [CrossRef]
- Solforosi, L.; Mancini, N.; Canducci, F.; Clementi, N.; Sautto, G.A.; Diotti, R.A.; Clementi, M.; Burioni, R. A phage display vector optimized for the generation of human antibody combinatorial libraries and the molecular cloning of monoclonal antibody fragments. New Microbiol. 2012, 35, 289–294. [Google Scholar]
- Lelli, D.; Moreno, A.; Brocchi, E.; Sozzi, E.; Capucci, L.; Canelli, E.; Barbieri, I.; Zeller, H.; Cordioli, P. West Nile virus: characterization and diagnostic applications of monoclonal antibodies. Virol. J. 2012, 9, 81. [Google Scholar] [CrossRef]
- Goodchild, S.A.; Dooley, H.; Schoepp, R.J.; Flajnik, M.; Lonsdale, S.G. Isolation and characterisation of Ebolavirus-specific recombinant antibody fragments from murine and shark immune libraries. Mol. Immunol. 2011, 48, 2027–2037. [Google Scholar] [CrossRef]
- De Genst, E.; Silence, K.; Decanniere, K.; Conrath, K.; Loris, R.; Kinne, J.; Muyldermans, S.; Wyns, L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. USA 2006, 103, 4586–4591. [Google Scholar]
- De Genst, E.; Saerens, D.; Muyldermans, S.; Conrath, K. Antibody repertoire development in camelids. Dev. Comp. Immunol. 2006, 30, 187–198. [Google Scholar] [CrossRef]
- Mancini, N.; Diotti, R.A.; Perotti, M.; Sautto, G.; Clementi, N.; Nitti, G.; Patel, A.H.; Ball, J.K.; Clementi, M.; Burioni, R. Hepatitis C virus (HCV) infection may elicit neutralizing antibodies targeting epitopes conserved in all viral genotypes. PloS One 2009, 4, e8254. [Google Scholar]
- Hu, H.; Voss, J.; Zhang, G.; Buchy, P.; Zuo, T.; Wang, L.; Wang, F.; Zhou, F.; Wang, G.; Tsai, C.; Calder, L.; Gamblin, S.J.; Zhang, L.; Deubel, V.; Zhou, B.; Skehel, J.J.; Zhou, P. A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J. Virol. 2012, 86, 2978–2989. [Google Scholar]
- Ohkura, T.; Kikuchi, Y.; Kono, N.; Itamura, S.; Komase, K.; Momose, F.; Morikawa, Y. Epitope mapping of neutralizing monoclonal antibody in avian influenza A H5N1 virus hemagglutinin. Biochem. Biophys. Res. Commun. 2012, 418, 38–43. [Google Scholar] [CrossRef]
- Liang, L.; Huang, P.; Wen, M.; Ni, H.; Tan, S.; Zhang, Y.; Chen, Q. Epitope peptides of influenza H3N2 virus neuraminidase gene designed by immunoinformatics. Acta Biochim. Biophys. Sin. (Shanghai) 2012, 44, 113–118. [Google Scholar] [CrossRef]
- Bugli, F.; Mancini, N.; Kang, C.Y.; Di Campli, C.; Grieco, A.; Manzin, A.; Gabrielli, A.; Gasbarrini, A.; Fadda, G.; Varaldo, P.E.; Clementi, M.; Burioni, R. Mapping B-cell epitopes of hepatitis C virus E2 glycoprotein using human monoclonal antibodies from phage display libraries. J. Virol. 2001, 75, 9986–9990. [Google Scholar]
- Sakabe, S.; Iwatsuki-Horimoto, K.; Horimoto, T.; Nidom, C.A.; Le, M.Q.; Takano, R.; Kubota-Koketsu, R.; Okuno, Y.; Ozawa, M.; Kawaoka, Y. A cross-reactive neutralizing monoclonal antibody protects mice from H5N1 and pandemic (H1N1) 2009 virus infection. Antiviral Res. 2010, 88, 249–255. [Google Scholar] [CrossRef]
- Serruto, D.; Adu-Bobie, J.; Capecchi, B.; Rappuoli, R.; Pizza, M.; Masignani, V. Biotechnology and vaccines: application of functional genomics to Neisseria meningitidis and other bacterial pathogens. J. Biotechnol. 2004, 113, 15–32. [Google Scholar]
- Cao, Z.; Meng, J.; Li, X.; Wu, R.; Huang, Y.; He, Y. The epitope and neutralization mechanism of AVFluIgG01, a broad-reactive human monoclonal antibody against H5N1 influenza virus. PloS One 2012, 7, e38126. [Google Scholar]
- Corallini, A.; Mazzoni, E.; Taronna, A.; Manfrini, M.; Carandina, G.; Guerra, G.; Guaschino, R.; Vaniglia, F.; Magnani, C.; Casali, F.; Dolcetti, R.; Palmonari, C.; Rezza, G.; Martini, F.; Barbanti-Brodano, G.; Tognon, M.G. Specific antibodies reacting with simian virus 40 capsid protein mimotopes in serum samples from healthy blood donors. Hum. Immun. 2012, 73, 502–510. [Google Scholar] [CrossRef]
- Wang, Y.S.; Ouyang, W.; Liu, X.J.; He, K.W.; Yu, S.Q.; Zhang, H.B.; Fan, H.J.; Lu, C.P. Virus-like particles of hepatitis B virus core protein containing five mimotopes of infectious bursal disease virus (IBDV) protect chickens against IBDV. Vaccine 2012, 30, 2125–2130. [Google Scholar]
- Hall, T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Group, W.O.F.H.N.E.W. Continued evolution of highly pathogenic avian influenza A (H5N1): updated nomenclature. Influenza and Other Respi. Viruses 2012, 6, 1–5. [Google Scholar] [CrossRef]
- WHO. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. Releve epidemiologique hebdomadaire / Section d'hygiene du Secretariat de la Societe des Nations (Weekly epidemiological record / Health Section of the Secretariat of the League of Nations) 2012, 87, 97–108.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Clementi, N.; Criscuolo, E.; Castelli, M.; Mancini, N.; Clementi, M.; Burioni, R. Influenza B-Cells Protective Epitope Characterization: A Passkey for the Rational Design of New Broad-Range Anti-Influenza Vaccines. Viruses 2012, 4, 3090-3108. https://doi.org/10.3390/v4113090
Clementi N, Criscuolo E, Castelli M, Mancini N, Clementi M, Burioni R. Influenza B-Cells Protective Epitope Characterization: A Passkey for the Rational Design of New Broad-Range Anti-Influenza Vaccines. Viruses. 2012; 4(11):3090-3108. https://doi.org/10.3390/v4113090
Chicago/Turabian StyleClementi, Nicola, Elena Criscuolo, Matteo Castelli, Nicasio Mancini, Massimo Clementi, and Roberto Burioni. 2012. "Influenza B-Cells Protective Epitope Characterization: A Passkey for the Rational Design of New Broad-Range Anti-Influenza Vaccines" Viruses 4, no. 11: 3090-3108. https://doi.org/10.3390/v4113090