Bacteriophages with the Ability to Degrade Uropathogenic Escherichia Coli Biofilms
Abstract
:1. Introduction
2. Results and Discussion
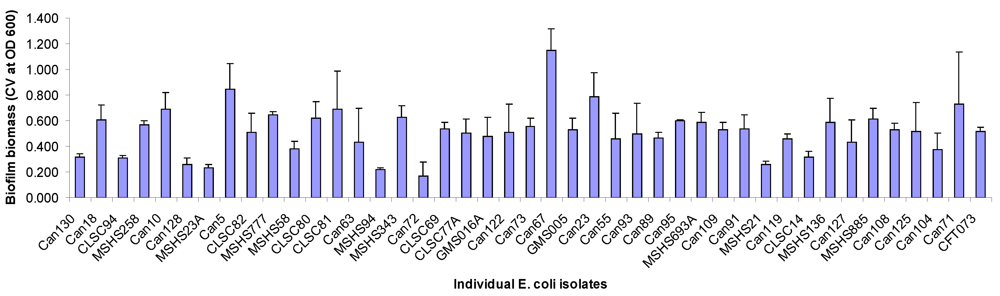
2.1. Biofilm Forming Capabilities, Serotype and Antimicrobial Resistance
| Strain | Serotype | Antimicrobials to which isolates are resistant | ACG-C40 | ACG-C91 | ACG-M12 |
|---|---|---|---|---|---|
| Can130 | O1:NM | + | + | - | |
| Can18 | O2:H4 | - | + | - | |
| CLSC94 | O2:H7 | + | + | - | |
| MSHS258 | O2:H18 | + | + | - | |
| Can10 | O8:H10 | - | - | - | |
| Can128 | O8:NM | SOX, TCY | + | - | - |
| MSHS23A | O6:H1 | AMP, STR, SOX, | + | - | + |
| Can5 | O6:H1 | NAL | + | - | + |
| CLSC82 | O6:H1 | AMP | + | - | + |
| MSHS777 | O6:H1 | STR | + | - | + |
| MSHS58 | O6:H25 | KAN | - | - | - |
| CLSC80 | O11:H18 | STR, SOX | + | - | - |
| CLSC81 | O14:H4 | + | - | + | |
| Can63 | O14:H31 | TCY | - | - | + |
| MSHS94 | O18ac:H7 | AMP, STR, SOX, TCY | + | + | + |
| MSHS343 | O18ac:H7 | + | + | + | |
| Can72 | O18ac:NM | AMP | + | - | - |
| CLSC69 | O21:H14 | - | - | - | |
| CLSC77A | O22:H1 | - | - | - | |
| GMS016A | O25:H1 | + | + | + | |
| Can122 | O25:H4 | AMP, TIO, CRO, CIP, NAL, SOX, TCY, SXT | - | - | - |
| Can73 | O25:H4 | AMP, CIP, NAL | - | - | - |
| Can67 | O25:H4 | AMC, AMP, CIP, NAL | - | - | - |
| GMS005A | O35:H10 | + | - | - | |
| Can23 | O68:H18 | + | - | - | |
| Can55 | O75:H7 | AMC, AMP, FOX, TIO, CRO | + | + | - |
| Can93 | O75:H7 | - | + | - | |
| Can89 | O78:H5 | + | - | - | |
| Can95 | O106:H18 | CHL, TCY | - | - | - |
| MSHS693A | O117:H5 | + | - | - | |
| Can109 | O134:H31 | NAL | + | + | - |
| Can91 | O135:H6 | + | + | + | |
| MSHS21 | O135:H11 | + | - | - | |
| Can119 | O153:H18 | AMP, TIO, CRO, NAL, STR, SOX, SXT | + | + | - |
| CLSC14 | O153:NM | AMP, STR, SOX, TCY, SXT | + | - | - |
| MSHS136 | O166:H15 | AMC, AMP | - | - | - |
| Can127 | OR:H4 | AMP, CHL, TCY | - | + | + |
| MSHS885 | OR:H4 | AMP, CHL, STR, SOX, TCY | + | + | - |
| Can108 | OR:H4 | SOX, TCY, STX | + | + | - |
| Can125 | OR:H4 | AMP, CIP, NAL, SOX, SXT | + | - | + |
| Can104 | OR:H7 | - | + | + | |
| Can71 | OR:H40 | + | - | + | |
| CFT073 | nd | nd | + | + | + |
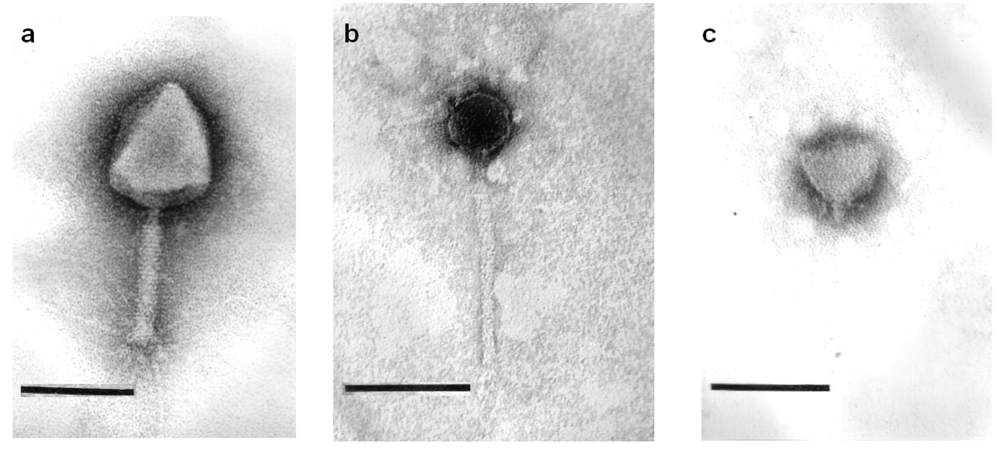
2.2. Phage Morphology
2.3. Lytic Spectra of Phages against Biofilm Forming UTI Isolates and Relation to Isolates’ Serotypes
2.4. Salient Bacteriophage Genome Features
2.4.1. Phage ACG-C40
2.4.2. Phage ACG-C91
2.4.3. Phage ACG-M12
2.5. Comparative Genomics
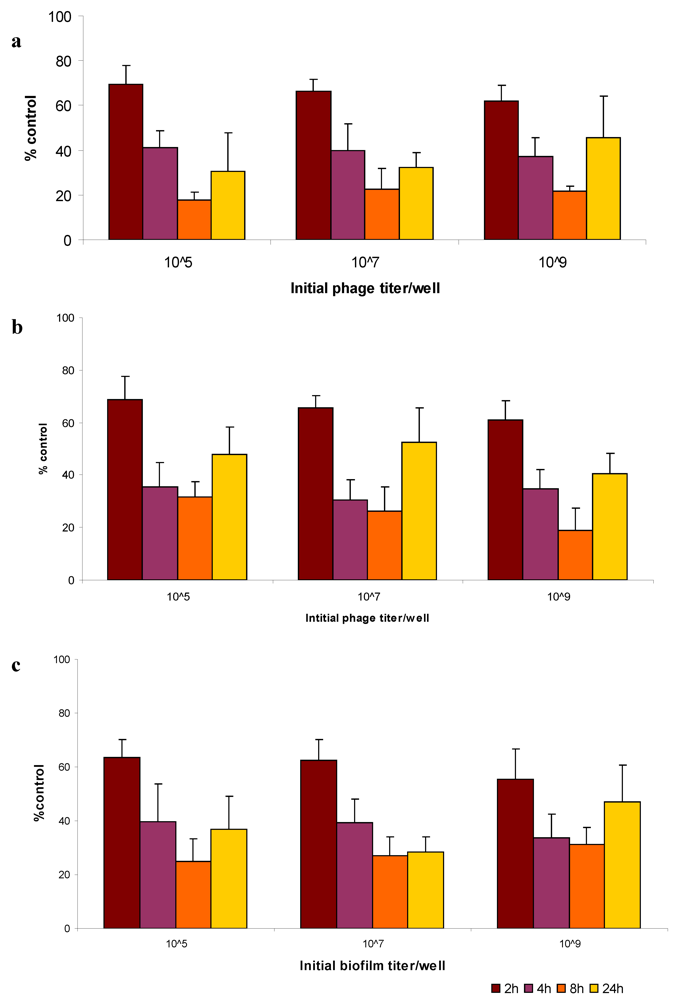
2.5. Bacteriophage Eradication of Established Biofilms
3. Experimental Section
3.1. Sampling and E. coli Strain Isolation
3.2. Culture Conditions
3.3. E. coli Serotyping
3.4. Susceptibility Testing
3.5. Bacteriophage Isolation
3.6. Transmission Electron Microscopy
3.7. Bacteriophage DNA Isolation, Restriction Analysis and Sequencing
3.8. Bioinformatic Analysis
3.9. Biofilm Assay
3.10. Bacteriophage Host Range on Biofilm Forming UPEC Isolates
3.11. Bacteriophage Activity against Established Biofilms
4. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Foxman, B. Epidemiology of urinary tract infections: Incidence, morbidity, and economic costs. Dis.-a-Mon. 2003, 49, 53–70. [Google Scholar] [CrossRef]
- Sanford, J.P. Urinary tract symptoms and infections. Ann. Rev. Med. 1975, 25, 485. [Google Scholar]
- Nicolle, L.E.; Ronald, A.R. Recurrent urinary tract infection in adult women: Diagnosis and treatment. Infect. Dis. Clin. N. Am. 1987, 1, 793–806. [Google Scholar]
- Nicolle, L.E. Epidemiology of urinary tract infections. Clin. Microbiol. Newslett. 2002, 24, 135–140. [Google Scholar]
- Czaja, C.A.; Hooton, T.M. Update on acute uncomplicated urinary tract infection in women. Postgrad. Med. 2006, 119, 39–45. [Google Scholar]
- Foxman, B.; Barlow, R.; D’Arcy, H.; Gillespie, B.; Sobel, J.D. Urinary tract infection: Self-reported incidence and associated costs. Ann. Epidemiol. 2000, 10, 509–515. [Google Scholar]
- Russo, T.A.; Johnson, J.R. Medical and economic impact of extraintestinal infections due to Escherichia coli: Focus on an increasingly important endemic problem. Microbes Infect. 2003, 5, 449–456. [Google Scholar] [CrossRef]
- Bacheller, C.D.; Bernstein, J.M. Urinary tract infections. Med. Clin. N. Am. 1997, 3, 719–730. [Google Scholar]
- Gupta, K.; Hooton, T.M.; Stamm, W.E. Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann. Intern. Med. 2001, 135, 41–50. [Google Scholar]
- Gupta, K. Addressing antibiotic resistance. Am. J. Med. 2002, 113, 29S–34S. [Google Scholar]
- Karlowsky, J.A.; Kelly, L.J.; Thornsberry, C.; Jones, M.E.; Sahm, D.F. Trends in antimicrobial resistance among urinary tract infection isolates of Escherichia coli from female outpatients in the United States. Antimicrob. Agents Chemother. 2002, 46, 2540–2545. [Google Scholar] [CrossRef]
- Karlowsky, J.A.; Thornsberry, C.; Jones, M.E.; Sahm, D.F. Susceptibility of antimicrobial-resistant urinary Escherichia coli isolates to fluoroquinolones and nitrofurantoin. Clin. Infect. Dis. 2003, 36, 183–187. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Hisanaga, T.L.; Laing, N.M.; DeCorby, M.R.; Nichol, K.A.; Palatnik, L.P.; Johnson, J.; Noreddin, A.; Harding, G.K.; Nicolle, L.E.; et al. NAUTICA Group. Antibiotic resistance in outpatient urinary isolates: Final results from the North American Urinary Tract Infection Collaborative Alliance (NAUTICA). Int. J. Antimicrob. Agents 2005, 26, 380–388. [Google Scholar]
- Morris, N.S.; Stickler, D.J.; McLean, R.J. The development of bacterial biofilms on indwelling urethral catheters. World J. Urol. 1999, 17, 345–350. [Google Scholar]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar]
- Doolittle, M.M.; Cooney, J.J.; Caldwell, D.E. Lytic infection of Escherichia coli biofilms by bacteriophage T4. Can. J. Microbiol. 1995, 42, 12–18. [Google Scholar]
- Doolittle, M.M.; Cooney, J.J.; Caldwell, D.E. Tracing the interaction of bacteriophage with bacterial biofilms using fluorescent and chromogenic probes. J. Indust. Microbiol. 1996, 16, 331–341. [Google Scholar]
- Lacroix-Gueu, P.; Briandet, R.; Leveque-Fort, S.; Bellon-Fontaine, M.N.; Fontaine-Aupart, M.P. In situ measurements of viral particles diffusion inside mucoid biofilms. C. R. Biol. 2005, 328, 1065–1072. [Google Scholar] [CrossRef]
- Wood, H.L.; Holden, S.R.; Bayston, R. Susceptibility of Staphylococcus epidermidis biofilm in CSF shunts to bacteriophage attack. Eur. J. Pediatr. Surg. 2001, 11, S56–S57. [Google Scholar]
- Sillankorva, S.; Oliveira, R.; Vieira, M.J.; Sutherland, I.W.; Azeredo, J. Bacteriophage φS1 infection of Pseudomonas fluorescens planktonic cells versus biofilms. Biofouling 2004, 20, 133–138. [Google Scholar] [CrossRef]
- Curtin, J.J.; Donlan, R.M. Using bacteriophages to reduce formation of catheter-associated biofilms by Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2006, 50, 1268–1275. [Google Scholar] [CrossRef]
- Hughes, K.A.; Sutherland, I.W.; Jones, M.V. Biofilm susceptibility to bacteriophage attack: The role of phage-borne polysaccharide depolymerise. Microbiology 1998, 144, 3039–3047. [Google Scholar]
- Hughes, K.A.; Sutherland, I.W.; Clark, J.; Jones, M.V. Bacteriophage and associated polysaccharide depolymerases-novel tools for study of bacterial biofilms. J. Appl. Microbiol. 1998, 85, 583–590. [Google Scholar]
- Hanlon, G.W.; Denyer, S.P.; Ollif, C.J.; Ibrahim, L.J. Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2001, 67, 2746–2753. [Google Scholar] [CrossRef]
- Rijavec, M.; Müller-Premru, M.; Zakotnik, B.; Zgur-Bertok, D. Virulence factors and biofilm production among Escherichia coli strains causing bacteraemia of urinary tract origin. J. Med. Microbiol. 2008, 57, 1329–1334. [Google Scholar] [CrossRef]
- Hoyle, B.D.; Alcantara, J.; Costerton, J.W. Pseudomonas aeruginosa biofilm as a diffusion barrier to piperacillin. Antimicrob.Agents Chemother. 1992, 36, 2054–2056. [Google Scholar]
- Ito, A.; Taniuchi, A.; May, T.; Kawata, K.; Okabe, S. Increased antibiotic resistance of Escherichia coli in mature biofilms. Appl. Environ. Microbiol. 2009, 75, 4093–4100. [Google Scholar] [CrossRef]
- Gilbert, P.; Maira-Litran, T.; McBain, A.J.; Rickard, A.H.; Whyte, F.W. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microbial. Physiol. 2002, 46, 203–256. [Google Scholar]
- Sillankorva, S.; Oliveira, D.; Moura, A.; Henriques, M.; Faustino, A.; Nicolau, A.; Azeredo, J. Efficacy of a broad host range lytic bacteriophage against E. coli adhered to urothelium. Curr. Microbiol. 2011, 62, 1128–1132. [Google Scholar] [CrossRef] [Green Version]
- Goodridge, L.; Gallaccio, A.; Griffiths, M.W. Morphological, host range, and genetic characterization of two coliphages. Appl. Environ. Microbiol. 2003, 69, 5364–5371. [Google Scholar] [CrossRef]
- Chibani-Chennoufi, S.; Sidoti, J.; Bruttin, A.; Dillmann, M.L.; Kutter, E.; Qadri, F.; Sarker, S.A.; Brüssow, H. Isolation of Escherichia coli bacteriophages from the stool of pediatric diarrhea patients in Bangladesh. J. Bacteriol. 2004, 186, 8287–8294. [Google Scholar] [CrossRef]
- Zuber, S.; Ngom-Bru, C.; Barretto, C.; Bruttin, A.; Brüssow, H.; Denou, E. Genome analysis of phage JS98 defines a fourth major subgroup of T4-like phages in Escherichia coli. J. Bacteriol. 2007, 189, 8206–8214. [Google Scholar] [CrossRef]
- Nishikawa, H.; Yasuda, M.; Uchiyama, J.; Rashel, M.; Maeda, Y.; Takemura, I.; Sugihara, S.; Ujihara, T.; Shimizu, Y.; Shuin, T.; et al. T-even-related bacteriophages as candidates for treatment of Escherichia coli urinary tract infections. Arch. Virol. 2008, 153, 507–515. [Google Scholar] [CrossRef]
- Verma, V.; Harjai, K.; Chibber, S. Structural changes induced by a lytic bacteriophage make ciprofloxacin effective against older biofilm of Klebsiella pneumoniae. Biofouling 2010, 26, 729–737. [Google Scholar] [CrossRef]
- Corbin, B.D.; McLean, R.J.; Aron, G.M. Bacteriophage T4 multiplication in a glucose-limited Escherichia coli biofilm. Can. J. Microbiol. 2001, 47, 680–684. [Google Scholar]
- Lu, T.K.; Collins, J.J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 11197–11202. [Google Scholar]
- Petrov, V.M.; Ratnayaka, S.; Nolan, J.M.; Miller, E.S.; Karam, J.D. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol. J. 2010, 7, 292. [Google Scholar]
- Carlson, K.; Raleigh, E.A.; Hattman, S. Restriction and Modification. In Bacteriophage T4; Karam, J.D., Ed.; ASM Press: Washington, DC, USA, 1994; pp. 369–381. [Google Scholar]
- Mahadevan, P.; King, J.F.; Seto, D. CGUG: In silico proteome and genome parsing tool for the determination of “core” and unique genes in the analysis of genomes up to ca. 1.9 Mb. BMC Res. Notes 2009, 2, 168. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar]
- Kropinski, A.M.; Borodovsky, M.; Carver, T.J.; Cerdeño-Tárraga, A.M.; Darling, A.; Lomsadze, A.; Mahadevan, P.; Stothard, P.; Seto, D.; Van Domselaar, G.; et al. In silico identification of genes in bacteriophage DNA. Methods Mol. Biol. 2009, 502, 57–89. [Google Scholar] [CrossRef]
- Canadian Antimicrobial Resistance Alliance. Available online: http://www.can-r.com/ (accessed on 26 March 2012).
- Zhanel, G.G.; Adam, H.J.; Low, D.E.; Blondeau, J.; Decorby, M.; Karlowsky, J.A.; Weshnoweski, B.; Vashisht, R.; Wierzbowski, A.; Hoban, D.J. Canadian antimicrobial resistance alliance (CARA). Antimicrobial susceptibility of 15,644 pathogens from Canadian hospitals: results of the CANWARD 2007-2009 study. Diagn. Microbiol. Infect. Dis. 2011, 69, 291–306. [Google Scholar] [CrossRef]
- Griffith, D.P.; Musher, D.M.; Itin, C. Urease: The primary cause of infection-induced urinary stones. Invest. Urol. 1976, 13, 346–350. [Google Scholar]
- Stickler, D.J.; Morris, N.S.; Winters, C. Simple physical model to study the formation and physiology of biofilms on urethral catheters. Methods Enzymol. 1999, 310, 494–501. [Google Scholar]
- Ewing, E.H. Genus Escherichia. In Identification of Enterobacteriaceae, 4th; Edwards, P.R., Ewing, W.H., Eds.; Elsevier Science: New York, NY, USA, 1986; pp. 96–134. [Google Scholar]
- Government of Canada, Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2005. Public Health Agency of Canada: Guelph, Ontario, Canada, 2007.
- Cockerill, F.R.; Wikler, M.A.; Bush, K. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement M100-S20; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2010. [Google Scholar]
- Casas, V.; Rohwer, F. Phage metagenomics. Methods Enzymol. 2007, 421, 259–268. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor: New York, NY, USA, 1989; Volume 2. [Google Scholar]
- Adams, M.H. Bacteriophages. Interscience Publishers: New York, NY, USA, 1959. [Google Scholar]
- Kropinski, A.M.; Prangishvili, D.; Lavigne, R. Position paper: The creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ. Microbiol. 2009, 11, 2775–2777. [Google Scholar]
- myRAST- The SEED Server. Available online: http://blog.theseed.org/servers/presentations/t1/drast-overview.html (accessed on 9 February 2012).
- GBKFAA. Available online: http://lfz.corefacility.ca/gbk2faa/ (accessed on 9 February 2012).
- Basic Local Alignment Search Tool (BLAST). Available online: http://blast.ncbi.nlm.nih.gov/ (accessed on 9 February 2012).
- Finn, R.D.; Mistry, J.; Tate, J.; Coggill, P.; Heger, A.; Pollington, J.E.; Gavin, O.L.; Gunasekaran, P.; Ceric, G.; Forslund, K.; et al. The Pfam protein families database. Nucleic Acids Res. 2010, 38, D211–D222. [Google Scholar]
- Sonnhammer, E.L.; Von, H.G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the International Conference on Intelligent Systems for Molecular Biology, Montreal, QC, Canada, 28 June–1 July 1998; 1998; pp. 175–182. [Google Scholar]
- Kall, L.; Krogh, A.; Sonnhammer, E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2011, 39, D32–D37. [Google Scholar]
- Genbank to Sequin File Converter. Available online: http://lfz.corefacility.ca/gbk2sqn/ (accessed on 9 February 2012).
- O’Toole, G.A.; Pratt, L.A.; Watnick, P.I.; Newman, D.K.; Weaver, V.B.; Kolter, R. Genetic approaches to study of biofilms. Methods Enzymol. 1999, 310, 91–109. [Google Scholar]
- Kutter, E. Phage Host Range and Efficiency of Plating. In Bacteriophages: Methods and Protocols; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: New York, NY, USA, 2009; Volume 1, pp. 141–151. [Google Scholar]
- Soni, K.A.; Nannapaneni, R. Removal of Listeria monocytogenes biofilms with bacteriophage P100. J. Food Prot. 2010, 73, 1519–1524. [Google Scholar]
Supplementary Files
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chibeu, A.; Lingohr, E.J.; Masson, L.; Manges, A.; Harel, J.; Ackermann, H.-W.; Kropinski, A.M.; Boerlin, P. Bacteriophages with the Ability to Degrade Uropathogenic Escherichia Coli Biofilms. Viruses 2012, 4, 471-487. https://doi.org/10.3390/v4040471
Chibeu A, Lingohr EJ, Masson L, Manges A, Harel J, Ackermann H-W, Kropinski AM, Boerlin P. Bacteriophages with the Ability to Degrade Uropathogenic Escherichia Coli Biofilms. Viruses. 2012; 4(4):471-487. https://doi.org/10.3390/v4040471
Chicago/Turabian StyleChibeu, Andrew, Erika J. Lingohr, Luke Masson, Amee Manges, Josée Harel, Hans-W. Ackermann, Andrew M. Kropinski, and Patrick Boerlin. 2012. "Bacteriophages with the Ability to Degrade Uropathogenic Escherichia Coli Biofilms" Viruses 4, no. 4: 471-487. https://doi.org/10.3390/v4040471




