RSV Fusion: Time for a New Model
Abstract
:1. Introduction
2. Viral Envelope Proteins and Fusion
3. Defining a Functional RSV Receptor
4. Nucleolin: A Functional Fusion Receptor of RSV [15]
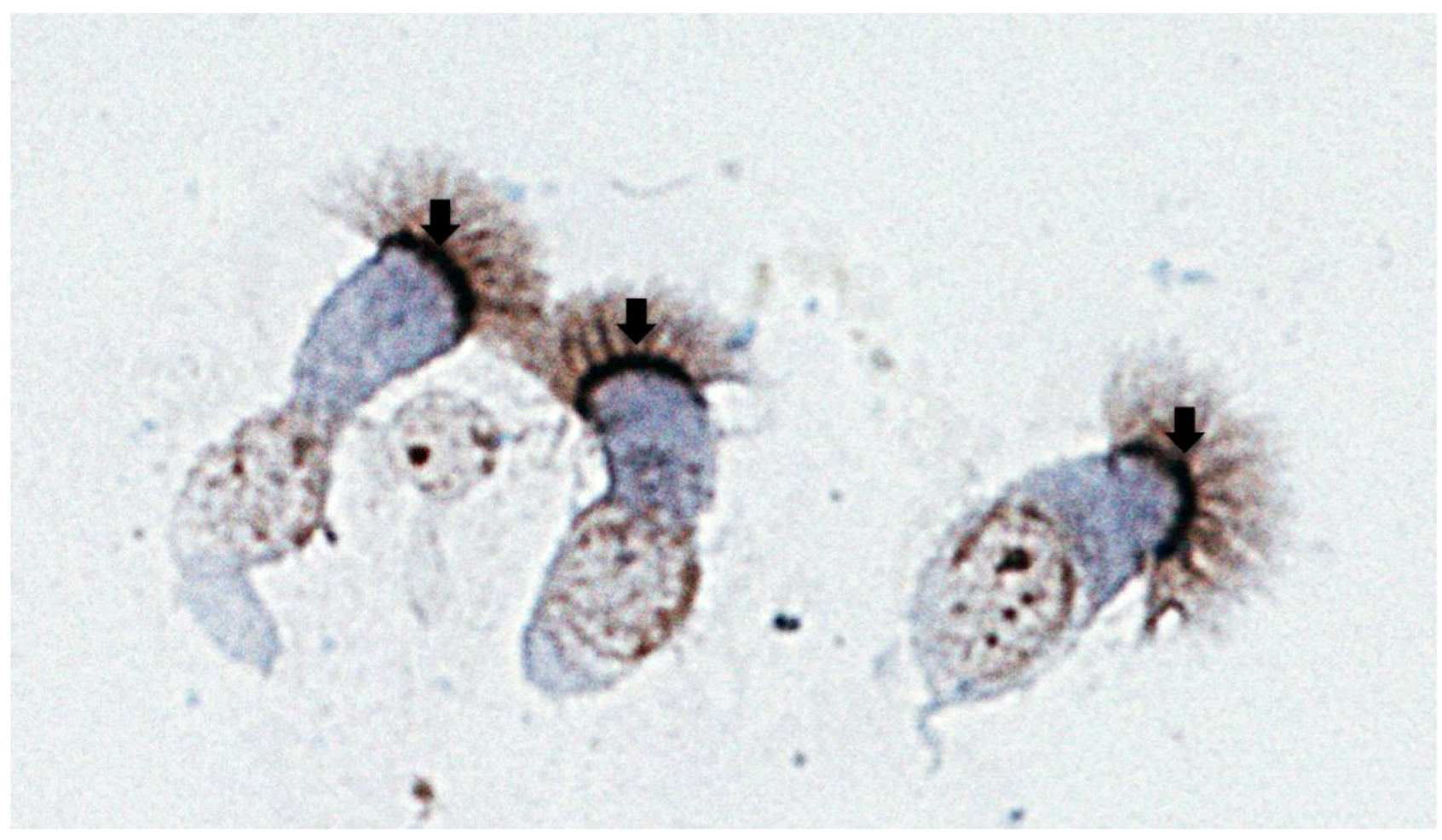
4.1. Identifying Nucleolin as a Ligand of Intact RSV
4.2. Nucleolin: Brief Overview
5. A New Model for RSV Fusion/Entry
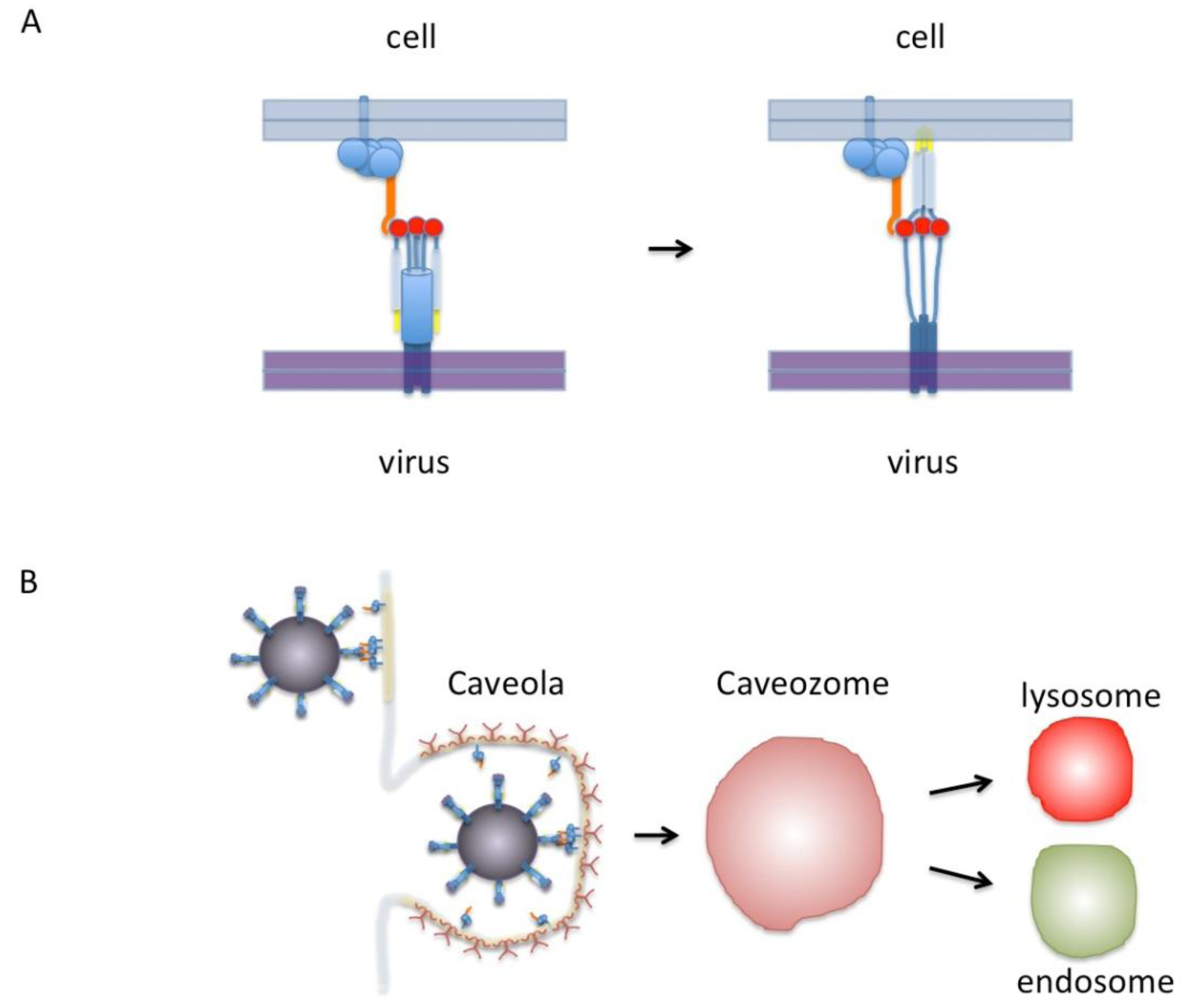
6. Future Directions
6.1. Targeting the Host in RSV Prophylaxis and Therapy?
6.2. Nucleolin as an Anti-RSV Target
6.3. RSV-Nucleolin Interaction Domain(s)
6.4. Nucleolin and RSV Tropism
7. Concluding Remarks
Conflict of Interest
References and Notes
- Nair, H.; Nokes, D.J.; Gessner, B.D.; Dherani, M.; Madhi, S.A.; Singleton, R.J.; O'Brien, K.L.; Roca, A.; Wright, P.F.; Bruce, N.; et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: A systematic review and meta-analysis. Lancet 2010, 375, 1545–1555. [Google Scholar] [CrossRef]
- Walsh, E.E. Respiratory syncytial virus infection in adults. Sem. Resp. Crit. Care M. 2011, 32, 423–432. [Google Scholar] [CrossRef]
- Blount, R.E., Jr.; Morris, J.A.; Savage, R.E. Recovery of cytopathogenic agent from chimpanzees with coryza. Proc. Soc. Exp. Biol. Med. 1956, 92, 544–549. [Google Scholar]
- Collins, P.L.; Melero, J.A. Progress in understanding and controlling respiratory syncytial virus: Still crazy after all these years. Virus Res. 2011, 162, 80–99. [Google Scholar]
- Techaarpornkul, S.; Barretto, N.; Peeples, M.E. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 2001, 75, 6825–6834. [Google Scholar] [CrossRef]
- Behera, A.K.; Matsuse, H.; Kumar, M.; Kong, X.; Lockey, R.F.; Mohapatra, S.S. Blocking intercellular adhesion molecule-1 on human epithelial cells decreases respiratory syncytial virus infection. Biochem. Biophys. Res. Commun. 2001, 280, 188–195. [Google Scholar] [CrossRef]
- Krusat, T.; Streckert, H.J. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch. Virol. 1997, 142, 1247–1254. [Google Scholar] [CrossRef]
- Malhotra, R.; Ward, M.; Bright, H.; Priest, R.; Foster, M.R.; Hurle, M.; Blair, E.; Bird, M. Isolation and characterisation of potential respiratory syncytial virus receptor(s) on epithelial cells. Microbes Infect. 2003, 5, 123–133. [Google Scholar] [CrossRef]
- Marr, N.; Turvey, S.E. Role of human TLR4 in respiratory syncytial virus-induced NF-kappaB activation, viral entry and replication. Innate Immun. 2012, 18, 856–865. [Google Scholar] [CrossRef]
- Harcourt, J.; Alvarez, R.; Jones, L.P.; Henderson, C.; Anderson, L.J.; Tripp, R.A. Respiratory syncytial virus G protein and G protein CX3C motif adversely affect CX3CR1+ T cell responses. J. Immunol. 2006, 176, 1600–1608. [Google Scholar]
- Dimmock, N.J.; Easton, A.J.; Leppard, K. Introduction to Modern Virology, 6th ed; Blackwell Pub.: Malden, MA, USA, 2007; p. 516. [Google Scholar]
- Duan, D.; Yue, Y.; Yan, Z.; McCray, P.B., Jr.; Engelhardt, J.F. Polarity influences the efficiency of recombinant adenoassociated virus infection in differentiated airway epithelia. Hum. Gene. Ther. 1998, 9, 2761–2776. [Google Scholar] [CrossRef]
- Techaarpornkul, S.; Collins, P.L.; Peeples, M.E. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology 2002, 294, 296–304. [Google Scholar] [CrossRef]
- Batonick, M.; Oomens, A.G.; Wertz, G.W. Human respiratory syncytial virus glycoproteins are not required for apical targeting and release from polarized epithelial cells. J. Virol. 2008, 82, 8664–8672. [Google Scholar] [CrossRef]
- Tayyari, F.; Marchant, D.; Moraes, T.J.; Duan, W.; Mastrangelo, P.; Hegele, R.G. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat. Med. 2011, 17, 1132–1135. [Google Scholar]
- Cao, W.; Henry, M.D.; Borrow, P.; Yamada, H.; Elder, J.H.; Ravkov, E.V.; Nichol, S.T.; Compans, R.W.; Campbell, K.P.; Oldstone, M.B. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 1998, 282, 2079–2081. [Google Scholar] [CrossRef]
- Reyes-Reyes, E.M.; Akiyama, S.K. Cell-surface nucleolin is a signal transducing P-selectin binding protein for human colon carcinoma cells. Exp. Cell. Res. 2008, 314, 2212–2223. [Google Scholar] [CrossRef]
- Losfeld, M.E.; Khoury, D.E.; Mariot, P.; Carpentier, M.; Krust, B.; Briand, J.P.; Mazurier, J.; Hovanessian, A.G.; Legrand, D. The cell surface expressed nucleolin is a glycoprotein that triggers calcium entry into mammalian cells. Exp. Cell. Res. 2009, 315, 357–369. [Google Scholar] [CrossRef]
- Orrick, L.R.; Olson, M.O.; Busch, H. Comparison of nucleolar proteins of normal rat liver and Novikoff hepatoma ascites cells by two-dimensional polyacrylamide gel electrophoresis. Proc. Natl. Acad. Sci. USA 1973, 70, 1316–1320. [Google Scholar] [CrossRef]
- Bugler, B.; Caizergues-Ferrer, M.; Bouche, G.; Bourbon, H.; Amalric, F. Detection and localization of a class of proteins immunologically related to a 100-kDa nucleolar protein. Eur. J. Biochem. 1982, 128, 475–480. [Google Scholar]
- Ginisty, H.; Sicard, H.; Roger, B.; Bouvet, P. Structure and functions of nucleolin. J. Cell Sci. 1999, 112, 761–772. [Google Scholar]
- Bicknell, K.; Brooks, G.; Kaiser, P.; Chen, H.; Dove, B.K.; Hiscox, J.A. Nucleolin is regulated both at the level of transcription and translation. Biochem. Biophys. Res. Commun. 2005, 332, 817–822. [Google Scholar] [CrossRef]
- Chen, C.M.; Chiang, S.Y.; Yeh, N.H. Increased stability of nucleolin in proliferating cells by inhibition of its self-cleaving activity. J. Biol. Chem. 1991, 266, 7754–7758. [Google Scholar]
- Srivastava, M.; Pollard, H.B. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 1999, 13, 1911–1922. [Google Scholar]
- Tajrishi, M.M.; Tuteja, R.; Tuteja, N. Nucleolin: The most abundant multifunctional phosphoprotein of nucleolus. Comm. Integ. Biol. 2011, 4, 267–275. [Google Scholar] [CrossRef]
- Hovanessian, A.G.; Soundaramourty, C.; El Khoury, D.; Nondier, I.; Svab, J.; Krust, B. Surface expressed nucleolin is constantly induced in tumor cells to mediate calcium-dependent ligand internalization. PLoS ONE 2010, 5, e15787. [Google Scholar]
- Schwab, M. S.; Dreyer, C. Protein phosphorylation sites regulate the function of the bipartite NLS of nucleolin. EJCB 1997, 73, 287–297. [Google Scholar]
- Hovanessian, A.G.; Puvion-Dutilleul, F.; Nisole, S.; Svab, J.; Perret, E.; Deng, J.S.; Krust, B. The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp. Cell. Res. 2000, 261, 312–328. [Google Scholar] [CrossRef]
- Krust, B.; El Khoury, D.; Nondier, I.; Soundaramourty, C.; Hovanessian, A.G. Targeting surface nucleolin with multivalent HB-19 and related Nucant pseudopeptides results in distinct inhibitory mechanisms depending on the malignant tumor cell type. BMC Cancer 2011, 11, 333. [Google Scholar] [CrossRef]
- Chen, X.; Kube, D.M.; Cooper, M.J.; Davis, P.B. Cell surface nucleolin serves as receptor for DNA nanoparticles composed of pegylated polylysine and DNA. Mol. Ther. 2008, 16, 333–342. [Google Scholar] [CrossRef]
- Barel, M.; Hovanessian, A. G.; Meibom, K.; Briand, J. P.; Dupuis, M.; Charbit, A. A novel receptor—ligand pathway for entry of Francisella tularensis in monocyte-like THP-1 cells: Interaction between surface nucleolin and bacterial elongation factor Tu. BMC microbiology 2008, 8, 145. [Google Scholar] [CrossRef]
- Sinclair, J.F.; Dean-Nystrom, E.A.; O'Brien, A.D. The established intimin receptor Tir and the putative eucaryotic intimin receptors nucleolin and beta1 integrin localize at or near the site of enterohemorrhagic Escherichia coli O157:H7 adherence to enterocytes in vivo. Infect. Immun. 2006, 74, 1255–1265. [Google Scholar] [CrossRef]
- Calle, A.; Ugrinova, I.; Epstein, A.L.; Bouvet, P.; Diaz, J.J.; Greco, A. Nucleolin is required for an efficient herpes simplex virus type 1 infection. J. Virol. 2008, 82, 4762–4773. [Google Scholar] [CrossRef]
- Greco, A.; Arata, L.; Soler, E.; Gaume, X.; Coute, Y.; Hacot, S.; Calle, A.; Monier, K.; Epstein, A.L.; Sanchez, J.C.; et al. Nucleolin interacts with US11 protein of herpes simplex virus 1 and is involved in its trafficking. J. Virol. 2012, 86, 1449–1457. [Google Scholar] [CrossRef]
- Kusakawa, T.; Shimakami, T.; Kaneko, S.; Yoshioka, K.; Murakami, S. Functional interaction of hepatitis C Virus NS5B with Nucleolin GAR domain. J. Biochem. 2007, 141, 917–927. [Google Scholar] [CrossRef]
- Strang, B.L.; Boulant, S.; Kirchhausen, T.; Coen, D.M. Host cell nucleolin is required to maintain the architecture of human cytomegalovirus replication compartments. mBio 2012, 3, e00301–11. [Google Scholar]
- Cancio-Lonches, C.; Yocupicio-Monroy, M.; Sandoval-Jaime, C.; Galvan-Mendoza, I.; Urena, L.; Vashist, S.; Goodfellow, I.; Salas-Benito, J.; Gutierrez-Escolano, A.L. Nucleolin interacts with the feline calicivirus 3' untranslated region and the protease-polymerase NS6 and NS7 proteins, playing a role in virus replication. J. Virol. 2011, 85, 8056–8068. [Google Scholar] [CrossRef]
- Chang, A.; Dutch, R.E. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses 2012, 4, 613–636. [Google Scholar] [CrossRef]
- Harrison, S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 690–698. [Google Scholar] [CrossRef]
- Nisole, S.; Krust, B.; Hovanessian, A.G. Anchorage of HIV on permissive cells leads to coaggregation of viral particles with surface nucleolin at membrane raft microdomains. Exp. Cell. Res. 2002, 276, 155–173. [Google Scholar] [CrossRef]
- Chang, T.H.; Segovia, J.; Sabbah, A.; Mgbemena, V.; Bose, S. Cholesterol-rich lipid rafts are required for release of infectious human respiratory syncytial virus particles. Virology 2012, 422, 205–213. [Google Scholar] [CrossRef]
- Chen, X.; Shank, S.; Davis, P.B.; Ziady, A.G. Nucleolin-mediated cellular trafficking of DNA nanoparticle is lipid raft and microtubule dependent and can be modulated by glucocorticoid. Mol. Ther. 2011, 19, 93–102. [Google Scholar] [CrossRef]
- Werling, D.; Hope, J.C.; Chaplin, P.; Collins, R.A.; Taylor, G.; Howard, C.J. Involvement of caveolae in the uptake of respiratory syncytial virus antigen by dendritic cells. J. Leukoc. Biol. 1999, 66, 50–58. [Google Scholar]
- White, J.M.; Delos, S.E.; Brecher, M.; Schornberg, K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 189–219. [Google Scholar] [CrossRef]
- Kahn, J.S.; Schnell, M.J.; Buonocore, L.; Rose, J.K. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (RSV) glycoproteins: RSV fusion protein can mediate infection and cell fusion. Virology 1999, 254, 81–91. [Google Scholar] [CrossRef]
- Srinivasakumar, N.; Ogra, P.L.; Flanagan, T.D. Characteristics of fusion of respiratory syncytial virus with HEp-2 cells as measured by R18 fluorescence dequenching assay. J. Virol. 1991, 65, 4063–4069. [Google Scholar]
- Shahrabadi, M.S.; Lee, P.W. Calcium requirement for syncytium formation in HEp-2 cells by respiratory syncytial virus. J. Clin. Microbiol. 1988, 26, 139–141. [Google Scholar]
- Kolokoltsov, A.A.; Deniger, D.; Fleming, E.H.; Roberts, N.J., Jr.; Karpilow, J.M.; Davey, R.A. Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. J. Virol. 2007, 81, 7786–7800. [Google Scholar] [CrossRef]
- Song, N.; Ding, Y.; Zhuo, W.; He, T.; Fu, Z.; Chen, Y.; Song, X.; Fu, Y.; Luo, Y. The nuclear translocation of endostatin is mediated by its receptor nucleolin in endothelial cells. Angiogenesis 2012, 15, 697–711. [Google Scholar] [CrossRef]
- Adams, O.; Bonzel, L.; Kovacevic, A.; Mayatepek, E.; Hoehn, T.; Vogel, M. Palivizumab-resistant human respiratory syncytial virus infection in infancy. Clin. Infect. Dis. 2010, 51, 185–188. [Google Scholar] [CrossRef]
- Deng, J.S.; Ballou, B.; Hofmeister, J.K. Internalization of anti-nucleolin antibody into viable HEp-2 cells. Mol. Biol. Rep. 1996, 23, 191–195. [Google Scholar] [CrossRef]
- Bates, P.J.; Laber, D.A.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009, 86, 151–164. [Google Scholar] [CrossRef]
- Taylor, G.; Stott, E.J.; Furze, J.; Ford, J.; Sopp, P. Protective epitopes on the fusion protein of respiratory syncytial virus recognized by murine and bovine monoclonal antibodies. J. Gen. Virol. 1992, 73, 2217–2223. [Google Scholar] [CrossRef]
- Arbiza, J.; Taylor, G.; Lopez, J.A.; Furze, J.; Wyld, S.; Whyte, P.; Stott, E.J.; Wertz, G.; Sullender, W.; Trudel, M.; et al. Characterization of two antigenic sites recognized by neutralizing monoclonal antibodies directed against the fusion glycoprotein of human respiratory syncytial virus. J. Gen. Virol. 1992, 73, 2225–2234. [Google Scholar] [CrossRef]
- McLellan, J.S.; Yang, Y.; Graham, B.S.; Kwong, P.D. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J. Virol. 2011, 85, 7788–7796. [Google Scholar] [CrossRef]
- Schneider-Schaulies, J. Cellular receptors for viruses: links to tropism and pathogenesis. J. Gen. Virol. 2000, 81, 1413–1429. [Google Scholar]
- Asher, L.V.; Binn, L.N.; Mensing, T.L.; Marchwicki, R.H.; Vassell, R.A.; Young, G.D. Pathogenesis of hepatitis A in orally inoculated owl monkeys (Aotus trivirgatus). J. Med. Virol. 1995, 47, 260–268. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mastrangelo, P.; Hegele, R.G. RSV Fusion: Time for a New Model. Viruses 2013, 5, 873-885. https://doi.org/10.3390/v5030873
Mastrangelo P, Hegele RG. RSV Fusion: Time for a New Model. Viruses. 2013; 5(3):873-885. https://doi.org/10.3390/v5030873
Chicago/Turabian StyleMastrangelo, Peter, and Richard G. Hegele. 2013. "RSV Fusion: Time for a New Model" Viruses 5, no. 3: 873-885. https://doi.org/10.3390/v5030873




