The Foamy Virus Gag Proteins: What Makes Them Different?
Abstract
:1. Introduction
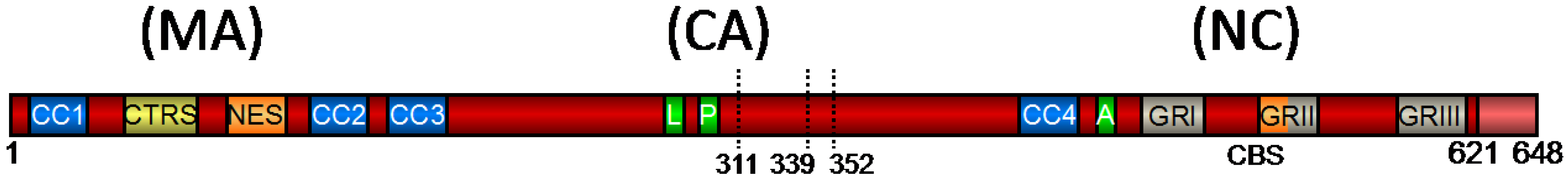
2. Functional Domains of PFV Gag and Their Role in Viral Replication
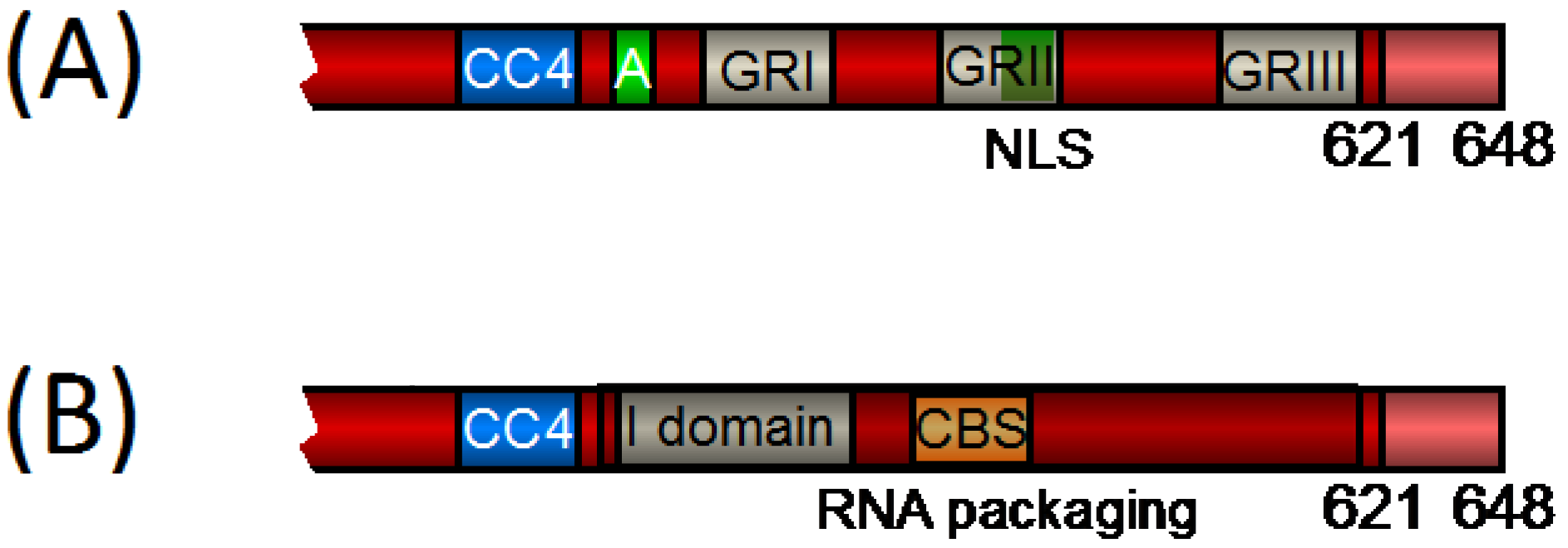
2.1. FV Gag Domains Found Also in Orthoretroviruses
2.4. Many Ways Lead to the Nucleus—FV Gag Nuclear Trafficking
| Viral entry | Nuclear translocation of the PIC | HIV‑1 (?), PFV (?) |
| Integration target site selection | MLV, HIV‑1, PFV (?) | |
| Chromatin tethering | PFV (?) | |
| Transcriptional regulation | Transcriptional enhancement of early gene expression | HIV‑1 |
| Regulation of cellular gene expression | (?) | |
| Viral assembly | Gag dimerization | RSV, PFV (?) |
| RNA binding/export | MLV, FIV, RSV, PFV (?) | |
| Pol binding/packaging | PFV (?) | |
| Other | Binding to translation machinery | MMTV |
| Intracellular retrotransposition | PFV (?) | |
| Regulation of splicing | MLV |
3. Interaction Between FV Gag and Host Cell Proteins
| Interactor | Function | Interaction Domain | Assay |
|---|---|---|---|
| Actin | Regulation of secondary Gag cleavage | Unknown | Co-immunoprecipitation |
| DyneinLC8 | Incoming capsid transport | CC3 | Co-immunoprecipitation, domain mutation |
| H2A/H2B | Chromatin tethering | CBS | Co-immunoprecipitation, domain mutation |
| CRM1 | Nuclear export | NES | Not shown |
| DDX6 | RNA encapsidation | Unknown | Co-localization |
| TSG101 | Viral budding | PSAP L domain | Y2H |
4. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Freed, E.O. Hiv–1 gag proteins: Diverse functions in the virus life cycle. Virology 1998, 251, 1–15. [Google Scholar]
- Meiering, C.D.; Linial, M.L. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 2001, 14, 165–176. [Google Scholar]
- Saib, A. Non–primate foamy viruses. Curr. Top. Microbiol. Immunol. 2003, 277, 197–211. [Google Scholar]
- Linial, M.L. Foamy viruses are unconventional retroviruses. J. Virol. 1999, 73, 1747–1755. [Google Scholar]
- Rethwilm, A. Foamy virus vectors: An awaited alternative to gammaretro– and lentiviral vectors. Curr. Gene Ther. 2007, 7, 261–271. [Google Scholar] [CrossRef]
- Bruss, V. Hepatitis b virus morphogenesis. World J. Gastroenterol. 2007, 13, 65–73. [Google Scholar]
- Achong, B.G.; Mansell, P.W.; Epstein, M.A.; Clifford, P. An unusual virus in cultures from a human nasopharyngeal carcinoma. J. Natl. Cancer Inst. 1971, 46, 299–307. [Google Scholar]
- Herchenroder, O.; Renne, R.; Loncar, D.; Cobb, E.K.; Murthy, K.K.; Schneider, J.; Mergia, A.; Luciw, P.A. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (sfvcpz): High homology to human foamy virus (hfv). Virology 1994, 201, 187–199. [Google Scholar] [CrossRef]
- Schweizer, M.; Turek, R.; Hahn, H.; Schliephake, A.; Netzer, K.O.; Eder, G.; Reinhardt, M.; Rethwilm, A.; Neumann–Haefelin, D. Markers of foamy virus infections in monkeys, apes, and accidentally infected humans: Appropriate testing fails to confirm suspected foamy virus prevalence in humans. AIDS Res. Hum. Retroviruses 1995, 11, 161–170. [Google Scholar]
- Enssle, J.; Jordan, I.; Mauer, B.; Rethwilm, A. Foamy virus reverse transcriptase is expressed independently from the gag protein. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 4137–4141. [Google Scholar]
- Lochelt, M.; Flugel, R.M. The human foamy virus pol gene is expressed as a pro–pol polyprotein and not as a gag–pol fusion protein. J. Virol. 1996, 70, 1033–1040. [Google Scholar]
- Yu, S.F.; Baldwin, D.N.; Gwynn, S.R.; Yendapalli, S.; Linial, M.L. Human foamy virus replication: A pathway distinct from that of retroviruses and hepadnaviruses. Science 1996, 271, 1579–1582. [Google Scholar]
- Giron, M.L.; Colas, S.; Wybier, J.; Rozain, F.; Emanoil–Ravier, R. Expression and maturation of human foamy virus gag precursor polypeptides. J. Virol. 1997, 71, 1635–1639. [Google Scholar]
- Pfrepper, K.I.; Lochelt, M.; Rackwitz, H.R.; Schnolzer, M.; Heid, H.; Flugel, R.M. Molecular characterization of proteolytic processing of the gag proteins of human spumavirus. J. Virol. 1999, 73, 7907–7911. [Google Scholar]
- Enssle, J.; Fischer, N.; Moebes, A.; Mauer, B.; Smola, U.; Rethwilm, A. Carboxy–terminal cleavage of the human foamy virus gag precursor molecule is an essential step in the viral life cycle. J. Virol. 1997, 71, 7312–7317. [Google Scholar]
- Zemba, M.; Wilk, T.; Rutten, T.; Wagner, A.; Flugel, R.M.; Lochelt, M. The carboxy–terminal p3gag domain of the human foamy virus gag precursor is required for efficient virus infectivity. Virology 1998, 247, 7–13. [Google Scholar] [CrossRef]
- Konvalinka, J.; Lochelt, M.; Zentgraf, H.; Flugel, R.M.; Krausslich, H.G. Active foamy virus proteinase is essential for virus infectivity but not for formation of a pol polyprotein. J. Virol. 1995, 69, 7264–7268. [Google Scholar]
- Cartellieri, M.; Herchenroder, O.; Rudolph, W.; Heinkelein, M.; Lindemann, D.; Zentgraf, H.; Rethwilm, A. N–terminal gag domain required for foamy virus particle assembly and export. J. Virol. 2005, 79, 12464–12476. [Google Scholar] [CrossRef]
- Wang, J.; Guo, H.Y.; Jia, R.; Xu, X.; Tan, J.; Geng, Y.Q.; Qiao, W.T. Preparation of bfv gag antiserum and preliminary study on cellular distribution of bfv. Virol. Sin. 2010, 25, 115–122. [Google Scholar] [CrossRef]
- Lehmann–Che, J.; Renault, N.; Giron, M.L.; Roingeard, P.; Clave, E.; Tobaly –Tapiero, J.; Bittoun, P.; Toubert, A.; de The, H.; Saib, A. Centrosomal latency of incoming foamy viruses in resting cells. PLoS Pathog. 2007, 3, e74. [Google Scholar] [CrossRef]
- Hutter, S.; Mullers, E.; Stanke, N.; Reh, J.; Lindemann, D. Prototype foamy virus protease activity is essential for intra–particle reverse transcription initiation but not absolutely required for uncoating upon host cell entry. J. Virol. 2013.
- Lindemann, D.; Rethwilm, A. Foamy virus biology and its application for vector development. Viruses 2011, 3, 561–585. [Google Scholar] [CrossRef]
- Swanstrom, R.; Wills, J.W. Synthesis, assembly, and processing of viral proteins. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor (NY), 1997. [Google Scholar]
- Geiselhart, V.; Schwantes, A.; Bastone, P.; Frech, M.; Lochelt, M. Features of the env leader protein and the n–terminal gag domain of feline foamy virus important for virus morphogenesis. Virology 2003, 310, 235–244. [Google Scholar]
- Life, R.B.; Lee, E.G.; Eastman, S.W.; Linial, M.L. Mutations in the amino terminus of foamy virus gag disrupt morphology and infectivity but do not target assembly. J. Virol. 2008, 82, 6109–6119. [Google Scholar] [CrossRef]
- Wilk, T.; Geiselhart, V.; Frech, M.; Fuller, S.D.; Flugel, R.M.; Lochelt, M. Specific interaction of a novel foamy virus env leader protein with the n–terminal gag domain. J. Virol. 2001, 75, 7995–8007. [Google Scholar] [CrossRef]
- Pietschmann, T.; Heinkelein, M.; Heldmann, M.; Zentgraf, H.; Rethwilm, A.; Lindemann, D. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 1999, 73, 2613–2621. [Google Scholar]
- Lindemann, D.; Pietschmann, T.; Picard–Maureau, M.; Berg, A.; Heinkelein, M.; Thurow, J.; Knaus, P.; Zentgraf, H.; Rethwilm, A. A particle–associated glycoprotein signal peptide essential for virus maturation and infectivity. 2001, 75, 5762–5771. [Google Scholar]
- Sfakianos, J.N.; Hunter, E. M–pmv capsid transport is mediated by env/gag interactions at the pericentriolar recycling endosome. Traffic 2003, 4, 671–680. [Google Scholar] [CrossRef]
- Yu, S.F.; Eastman, S.W.; Linial, M.L. Foamy virus capsid assembly occurs at a pericentriolar region through a cytoplasmic targeting/retention signal in gag. Traffic 2006, 7, 966–977. [Google Scholar] [CrossRef]
- Linial, M.L.; Eastman, S.W. Particle assembly and genome packaging. Curr. Top. Microbiol. Immunol. 2003, 277, 89–110. [Google Scholar]
- Eastman, S.W.; Linial, M.L. Identification of a conserved residue of foamy virus gag required for intracellular capsid assembly. J. Virol. 2001, 75, 6857–6864. [Google Scholar] [CrossRef]
- Choi, G.; Park, S.; Choi, B.; Hong, S.; Lee, J.; Hunter, E.; Rhee, S.S. Identification of a cytoplasmic targeting/retention signal in a retroviral gag polyprotein. J. Virol. 1999, 73, 5431–5437. [Google Scholar]
- Craven, R.C.; Parent, L.J. Dynamic interactions of the gag polyprotein. Curr. Top. Microbiol. Immunol. 1996, 214, 65–94. [Google Scholar]
- Tobaly–Tapiero, J.; Bittoun, P.; Giron, M.L.; Neves, M.; Koken, M.; Saib, A.; de The, H. Human foamy virus capsid formation requires an interaction domain in the n terminus of gag. J. Virol. 2001, 75, 4367–4375. [Google Scholar]
- Bowzard, J.B.; Bennett, R.P.; Krishna, N.K.; Ernst, S.M.; Rein, A.; Wills, J.W. Importance of basic residues in the nucleocapsid sequence for retrovirus gag assembly and complementation rescue. J. Virol. 1998, 72, 9034–9044. [Google Scholar]
- Mannigel, I.; Stange, A.; Zentgraf, H.; Lindemann, D. Correct capsid assembly mediated by a conserved yxxlgl motif in prototype foamy virus gag is essential for infectivity and reverse transcription of the viral genome. J. Virol. 2007, 81, 3317–3326. [Google Scholar] [CrossRef]
- Mullers, E.; Uhlig, T.; Stirnnagel, K.; Fiebig, U.; Zentgraf, H.; Lindemann, D. Novel functions of prototype foamy virus gag glycine– arginine–rich boxes in reverse transcription and particle morphogenesis. J. Virol. 2011, 85, 1452–1463. [Google Scholar] [CrossRef]
- Yu, S.F.; Edelmann, K.; Strong, R.K.; Moebes, A.; Rethwilm, A.; Linial, M.L. The carboxyl terminus of the human foamy virus gag protein contains separable nucleic acid binding and nuclear transport domains. J. Virol. 1996, 70, 8255–8262. [Google Scholar]
- Cimarelli, A.; Luban, J. Human immunodeficiency virus type 1 virion density is not determined by nucleocapsid basic residues. J. Virol. 2000, 74, 6734–6740. [Google Scholar] [CrossRef]
- Stange, A.; Mannigel, I.; Peters, K.; Heinkelein, M.; Stanke, N.; Cartellieri, M.; Gottlinger, H.; Rethwilm, A.; Zentgraf, H.; Lindemann, D. Characterization of prototype foamy virus gag late assembly domain motifs and their role in particle egress and infectivity. J. Virol. 2005, 79, 5466–5476. [Google Scholar] [CrossRef]
- Patton, G.S.; Morris, S.A.; Chung, W.; Bieniasz, P.D.; McClure, M.O. Identification of domains in gag important for prototypic foamy virus egress. J. Virol. 2005, 79, 6392–6399. [Google Scholar] [CrossRef]
- Pincetic, A.; Leis, J. The mechanism of budding of retroviruses from cell membranes. Adv. Virol. 2009, 2009, 6239691–6239699. [Google Scholar]
- Pornillos, O.; Garrus, J.E.; Sundquist, W.I. Mechanisms of enveloped rna virus budding. Trends Cell. Biol. 2002, 12, 569–579. [Google Scholar] [CrossRef]
- Morita, E.; Sundquist, W.I. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004, 20, 395–425. [Google Scholar] [CrossRef]
- Thumer, L.; Rethwilm, A.; Holmes, E.C.; Bodem, J. The complete nucleotide sequence of a new world simian foamy virus. Virology 2007, 369, 191–197. [Google Scholar] [CrossRef]
- Maurer, B.; Flugel, R.M. Genomic organization of the human spumaretrovirus and its relatedness to aids and other retroviruses. AIDS Res. Hum. 1988, 4, 467–473. [Google Scholar] [CrossRef]
- Matthes, D.; Wiktorowicz, T.; Zahn, J.; Bodem, J.; Stanke, N.; Lindemann, D.; Rethwilm, A. Basic residues in the foamy virus gag protein. J. Virol. 2011, 85, 3986–3995. [Google Scholar] [CrossRef]
- Fischer, N.; Heinkelein, M.; Lindemann, D.; Enssle, J.; Baum, C.; Werder, E.; Zentgraf, H.; Muller, J.G.; Rethwilm, A. Foamy virus particle formation. J. Virol. 1998, 72, 1610–1615. [Google Scholar]
- Lindemann, D.; Goepfert, P.A. The foamy virus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 2003, 277, 111–129. [Google Scholar]
- Zhadina, M.; McClure, M.O.; Johnson, M.C.; Bieniasz, P.D. Ubiquitin–dependent virus particle budding without viral protein ubiquitination. Proc. Natl. Acad. Sci. USA 2007, 104, 20031–20036. [Google Scholar]
- Liu, Y.; Kim, Y.B.; Lochelt, M. N–terminally myristoylated feline foamy virus gag allows env–independent budding of sub–viral particles. Viruses 2011, 3, 2223–2237. [Google Scholar] [CrossRef]
- Swiersy, A.; Wiek, C.; Zentgraf, H.; Lindemann, D. Characterization and manipulation of foamy virus membrane interactions. Cell. Microbiol. 2012.
- Wills, J.W.; Craven, R.C. Form, function, and use of retroviral gag proteins. Aids 1991, 5, 639–654. [Google Scholar] [CrossRef]
- Dorfman, T.; Luban, J.; Goff, S.P.; Haseltine, W.A.; Gottlinger, H.G. Mapping of functionally important residues of a cysteine–histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1993, 67, 6159–6169. [Google Scholar]
- Aldovini, A.; Young, R.A. Mutations of rna and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 1990, 64, 1920–1926. [Google Scholar]
- D'Souza, V.; Summers, M.F. How retroviruses select their genomes. Nature Rev. Microbiol. 2005, 3, 643–655. [Google Scholar] [CrossRef]
- Schwartz, M.D.; Fiore, D.; Panganiban, A.T. Distinct functions and requirements for the cys–his boxes of the human immunodeficiency virus type 1 nucleocapsid protein during rna encapsidation and replication. J. Virol. 1997, 71, 9295–9305. [Google Scholar]
- Winkler, I.; Bodem, J.; Haas, L.; Zemba, M.; Delius, H.; Flower, R.; Flugel, R.M.; Lochelt, M. Characterization of the genome of feline foamy virus and its proteins shows distinct features different from those of primate spumaviruses. J. Virol. 1997, 71, 6727–6741. [Google Scholar]
- Gabus, C.; Ivanyi–Nagy, R.; Depollier, J.; Bucheton, A.; Pelisson, A.; Darlix, J.L. Characterization of a nucleocapsid–like region and of two distinct primer trnalys,2 binding sites in the endogenous retrovirus gypsy. Nucleic Acids Res. 2006, 34, 5764–5777. [Google Scholar] [CrossRef]
- Schliephake, A.W.; Rethwilm, A. Nuclear localization of foamy virus gag precursor protein. J. Virol. 1994, 68, 4946–4954. [Google Scholar]
- Tobaly–Tapiero, J.; Bittoun, P.; Neves, M.; Guillemin, M.C.; Lecellier, C.H.; Puvion–Dutilleul, F.; Gicquel, B.; Zientara, S.; Giron, M.L.; de The, H.; et al. Isolation and characterization of an equine foamy virus. J. Virol. 2000, 74, 4064–4073. [Google Scholar] [CrossRef]
- Hatton, T.; Zhou, S.; Standring, D.N. Rna– and DNA–binding activities in hepatitis b virus capsid protein: A model for their roles in viral replication. J. Virol. 1992, 66, 5232–5241. [Google Scholar]
- Stenbak, C.R.; Linial, M.L. Role of the c terminus of foamy virus gag in rna packaging and pol expression. J. Virol. 2004, 78, 9423–9430. [Google Scholar] [CrossRef]
- Heinkelein, M.; Leurs, C.; Rammling, M.; Peters, K.; Hanenberg, H.; Rethwilm, A. Pregenomic rna is required for efficient incorporation of pol polyprotein into foamy virus capsids. J. Virol. 2002, 76, 10069–10073. [Google Scholar] [CrossRef]
- Peters, K.; Wiktorowicz, T.; Heinkelein, M.; Rethwilm, A. Rna and protein requirements for incorporation of the pol protein into foamy virus particles. J. Virol. 2005, 79, 7005–7013. [Google Scholar] [CrossRef]
- Lee, E.G.; Linial, M.L. The c terminus of foamy retrovirus gag contains determinants for encapsidation of pol protein into virions. J. Virol. 2008, 82, 10803–10810. [Google Scholar] [CrossRef]
- Lee, E.G.; Linial, M.L. Deletion of a cys–his motif from the alpharetrovirus nucleocapsid domain reveals late domain mutant–like budding defects. Virology 2006, 347, 226–233. [Google Scholar] [CrossRef]
- Muriaux, D.; Costes, S.; Nagashima, K.; Mirro, J.; Cho, E.; Lockett, S.; Rein, A. Role of murine leukemia virus nucleocapsid protein in virus assembly. J. Virol. 2004, 78, 12378–12385. [Google Scholar] [CrossRef]
- Hooks, J.J.; Gibbs, C.J., Jr. The foamy viruses. Bacteriol. Rev. 1975, 39, 169–185. [Google Scholar]
- Dupont, S.; Sharova, N.; DeHoratius, C.; Virbasius, C.M.; Zhu, X.; Bukrinskaya, A.G.; Stevenson, M.; Green, M.R. A novel nuclear export activity in hiv–1 matrix protein required for viral replication. Nature 1999, 402, 681–685. [Google Scholar]
- Nash, M.A.; Meyer, M.K.; Decker, G.L.; Arlinghaus, R.B. A subset of pr65gag is nucleus associated in murine leukemia virus–infected cells. J. Virol. 1993, 67, 1350–1356. [Google Scholar]
- Scheifele, L.Z.; Garbitt, R.A.; Rhoads, J.D.; Parent, L.J. Nuclear entry and crm1–dependent nuclear export of the rous sarcoma virus gag polyprotein. Proc. Natl. Acad. Sci. USA 2002, 99, 3944–3949. [Google Scholar]
- Beyer, A.R.; Bann, D.V.; Rice, B.; Pultz, I.S.; Kane, M.; Goff, S.P.; Golovkina, T.V.; Parent, L.J. Nucleolar trafficking of the mouse mammary tumor virus gag protein induced by interaction with ribosomal protein l9. J. Virol. 2013, 87, 1069–1082. [Google Scholar] [CrossRef]
- Kong, X.H.; Yu, H.; Xuan, C.H.; Wang, J.Z.; Chen, Q.M.; Geng, Y.Q. The requirements and mechanism for capsid assembly and budding of bovine foamy virus. Arch. Virol. 2005, 150, 1677–1684. [Google Scholar] [CrossRef]
- Bodem, J.; Zemba, M.; Flugel, R.M. Nuclear localization of the functional bel 1 transactivator but not of the gag proteins of the feline foamy virus. Virology 1998, 251, 22–27. [Google Scholar] [CrossRef]
- Mullers, E.; Stirnnagel, K.; Kaulfuss, S.; Lindemann, D. Prototype foamy virus gag nuclear localization: A novel pathway among retroviruses. J. Virol. 2011, 85, 9276–9285. [Google Scholar] [CrossRef]
- Tobaly–Tapiero, J.; Bittoun, P.; Lehmann–Che, J.; Delelis, O.; Giron, M.L.; de The, H.; Saib, A. Chromatin tethering of incoming foamy virus by the structural gag protein. Traffic 2008, 9, 1717–1727. [Google Scholar]
- Stirnnagel, K.; Schupp, D.; Dupont, A.; Kudryavtsev, V.; Reh, J.; Mullers, E.; Lamb, D.C.; Lindemann, D. Differential ph–dependent cellular uptake pathways among foamy viruses elucidated using dual–colored fluorescent particles. Retrovirology 2012, 9, 71. [Google Scholar] [CrossRef]
- Renault, N.; Tobaly–Tapiero, J.; Paris, J.; Giron, M.L.; Coiffic, A.; Roingeard, P.; Saib, A. A nuclear export signal within the structural gag protein is required for prototype foamy virus replication. Retrovirology 2011, 8, 6. [Google Scholar] [CrossRef] [Green Version]
- Lo, Y.T.; Tian, T.; Nadeau, P.E.; Park, J.; Mergia, A. The foamy virus genome remains unintegrated in the nuclei of g1/s phase–arrested cells, and integrase is critical for preintegration complex transport into the nucleus. J. Virol. 2010, 84, 2832–2842. [Google Scholar] [CrossRef]
- Yamashita, M.; Emerman, M. Retroviral infection of non–dividing cells: Old and new perspectives. Virology 2006, 344, 88–93. [Google Scholar] [CrossRef]
- Heinkelein, M.; Pietschmann, T.; Jarmy, G.; Dressler, M.; Imrich, H.; Thurow, J.; Lindemann, D.; Bock, M.; Moebes, A.; Roy, J.; et al. Efficient intracellular retrotransposition of an exogenous primate retrovirus genome. EMBO J. 2000, 19, 3436–3445. [Google Scholar] [CrossRef]
- Roy, J.; Rudolph, W.; Juretzek, T.; Gartner, K.; Bock, M.; Herchenroder, O.; Lindemann, D.; Heinkelein, M.; Rethwilm, A. Feline foamy virus genome and replication strategy. J. Virol. 2003, 77, 11324–11331. [Google Scholar] [CrossRef]
- Kenney, S.P.; Lochmann, T.L.; Schmid, C.L.; Parent, L.J. Intermolecular interactions between retroviral gag proteins in the nucleus. J. Virol. 2008, 82, 683–691. [Google Scholar] [CrossRef]
- Parent, L.J. New insights into the nuclear localization of retroviral gag proteins. Nucleus 2011, 2, 92–97. [Google Scholar] [CrossRef]
- Morozov, V.A.; Copeland, T.D.; Nagashima, K.; Gonda, M.A.; Oroszlan, S. Protein composition and morphology of human foamy virus intracellular cores and extracellular particles. Virology 1997, 228, 307–317. [Google Scholar] [CrossRef]
- Bodem, J.; Schied, T.; Gabriel, R.; Rammling, M.; Rethwilm, A. Foamy virus nuclear rna export is distinct from that of other retroviruses. J. Virol. 2011, 85, 2333–2341. [Google Scholar]
- Pinney, J.W.; Dickerson, J.E.; Fu, W.; Sanders–Beer, B.E.; Ptak, R.G.; Robertson, D.L. Hiv–host interactions: A map of viral perturbation of the host system. Aids 2009, 23, 549–554. [Google Scholar]
- Ptak, R.G.; Fu, W.; Sanders–Beer, B.E.; Dickerson, J.E.; Pinney, J.W.; Robertson, D.L.; Rozanov, M.N.; Katz, K.S.; Maglott, D.R.; Pruitt, K.D.; et al. Cataloguing the hiv type 1 human protein interaction network. AIDS Res. Hum. Retroviruses 2008, 24, 1497–1502. [Google Scholar]
- Sastri, J.; Campbell, E.M. Recent insights into the mechanism and consequences of trim5alpha retroviral restriction. AIDS Res. Hum. Retroviruses 2011, 27, 231–238. [Google Scholar] [CrossRef]
- Bieniasz, P.D. Intrinsic immunity: A front–line defense against viral attack. Nat. Immunol. 2004, 5, 1109–1115. [Google Scholar] [CrossRef]
- Yap, M.W.; Lindemann, D.; Stanke, N.; Reh, J.; Westphal, D.; Hanenberg, H.; Ohkura, S.; Stoye, J.P. Restriction of foamy viruses by primate trim5alpha. J. Virol. 2008, 82, 5429–5439. [Google Scholar]
- Petit, C.; Giron, M.L.; Tobaly–Tapiero, J.; Bittoun, P.; Real, E.; Jacob, Y.; Tordo, N.; De The, H.; Saib, A. Targeting of incoming retroviral gag to the centrosome involves a direct interaction with the dynein light chain 8. J. Cell Sci. 2003, 116, 3433–3442. [Google Scholar] [CrossRef]
- Afonso, P.V.; Zamborlini, A.; Saib, A.; Mahieux, R. Centrosome and retroviruses: The dangerous liaisons. Retrovirology 2007, 4, 27. [Google Scholar] [CrossRef]
- Kutay, U.; Guttinger, S. Leucine–rich nuclear–export signals: Born to be weak. Trends Cell Biol. 2005, 15, 121–124. [Google Scholar] [CrossRef]
- Yu, S.F.; Lujan, P.; Jackson, D.L.; Emerman, M.; Linial, M.L. The dead–box rna helicase ddx6 is required for efficient encapsidation of a retroviral genome. PLoS Pathog. 2011, 7, e1002303. [Google Scholar] [CrossRef]
- Erlwein, O.; McClure, M. Gene delivery the key to gene therapy: The case for foamy viruses. Ther. Deliv. 2011, 2, 681–684. [Google Scholar] [CrossRef]
- Trobridge, G.D.; Horn, P.A.; Beard, B.C.; Kiem, H.P. Large animal models for foamy virus vector gene therapy. Viruses 2012, 4, 3572–3588. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Müllers, E. The Foamy Virus Gag Proteins: What Makes Them Different? Viruses 2013, 5, 1023-1041. https://doi.org/10.3390/v5041023
Müllers E. The Foamy Virus Gag Proteins: What Makes Them Different? Viruses. 2013; 5(4):1023-1041. https://doi.org/10.3390/v5041023
Chicago/Turabian StyleMüllers, Erik. 2013. "The Foamy Virus Gag Proteins: What Makes Them Different?" Viruses 5, no. 4: 1023-1041. https://doi.org/10.3390/v5041023