Suppression of Coronavirus Replication by Cyclophilin Inhibitors
Abstract
:1. Introduction
2. CsA Inhibits the Replication of Feline CoV
3. CsA Inhibits the Replication of Diverse CoVs
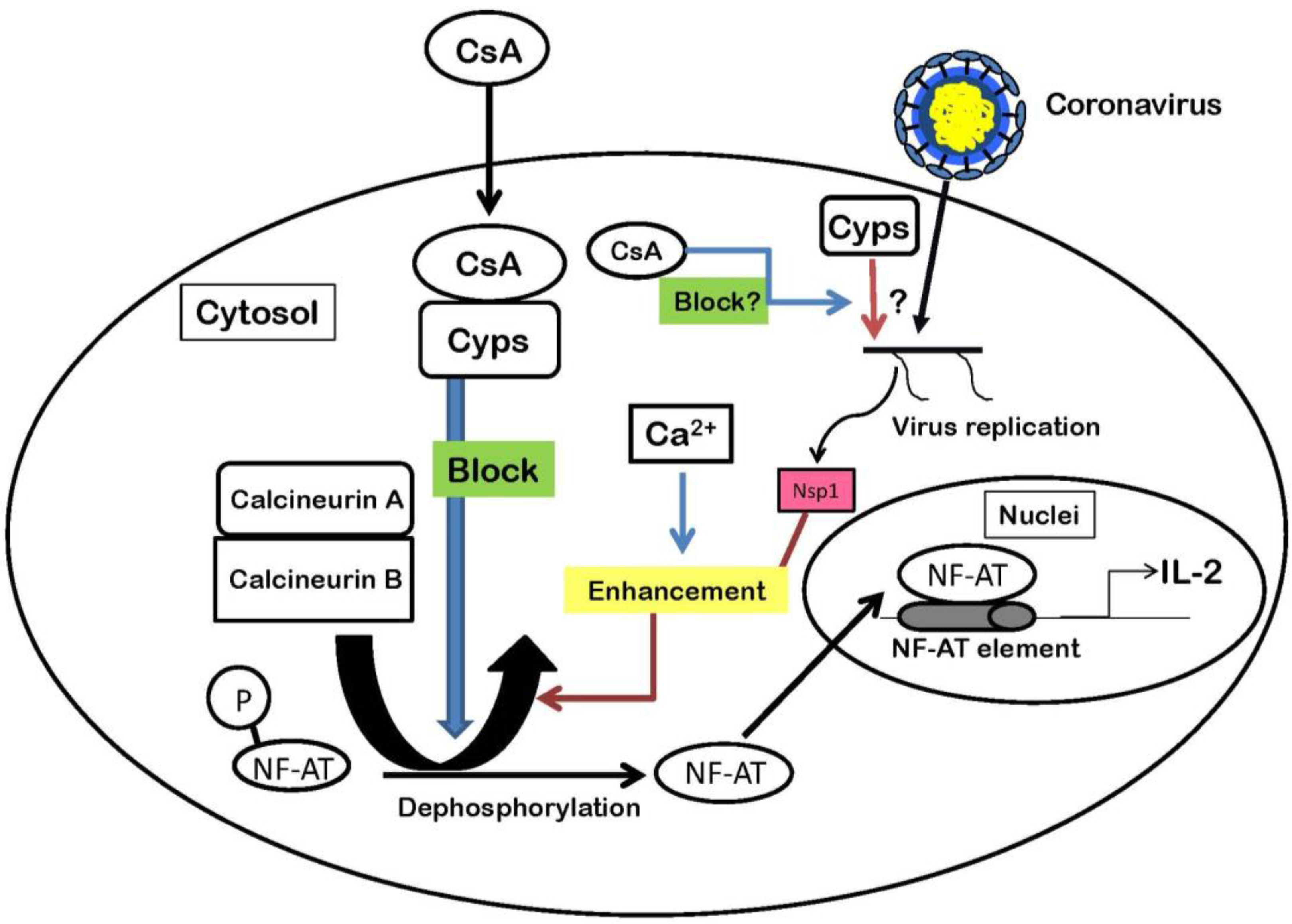
4. Nsp1 Protein Increases Signaling Through the Calcineurin/NF-AT Pathway
5. Cyp Inhibitors Block Arterivirus Replication
6. EAV Replication Depends on CypA
7. Conclusions
| Inhibition of virus replication by CsA treatment | Knockdown CypA | Knockdown CypB | Ref. | |||
|---|---|---|---|---|---|---|
| Virus replication | ||||||
| Nidovirales | Coronaviridae | SARS CoV | + | Not affected | Not affected | [25] |
| HCoV–NL63 | + | Unknown | Unknown | [26] | ||
| HCoV–229E | + | Unknown | Unknown | [26] | ||
| TGEV | + | Unknown | Unknown | [26] | ||
| IBV | + | Unknown | Unknown | [26] | ||
| MHV | + | Unknown | Unknown | [25] | ||
| FCoV | + | Unknown | Unknown | [26] | ||
| Arteriviridae | EAV | + | Inhibition of virus replication | Not affected | [33] | |
| PRRSV | + | Unknown | Unknown | [33] | ||
Acknowledgements
Conflict of Interest
References and Notes
- Perlman, S.; Netland, J. Coronaviruses post-sars: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef]
- Namy, O.; Moran, S.J.; Stuart, D.I.; Gilbert, R.J.; Brierley, I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 2006, 441, 244–247. [Google Scholar] [CrossRef]
- Sawicki, S.G.; Sawicki, D.L.; Siddell, S.G. A contemporary view of coronavirus transcription. J. Virol. 2007, 81, 20–29. [Google Scholar] [CrossRef]
- Quesniaux, V.F.; Schreier, M.H.; Wenger, R.M.; Hiestand, P.C.; Harding, M.W.; van Regenmortel, M.H. Cyclophilin binds to the region of cyclosporine involved in its immunosuppressive activity. Eur. J. Immunol. 1987, 17, 1359–1365. [Google Scholar] [CrossRef]
- Wang, P.; Heitman, J. The cyclophilins. Genome Biol. 2005, 6, e226. [Google Scholar] [CrossRef] [Green Version]
- Gething, M.J.; Sambrook, J. Protein folding in the cell. Nature 1992, 355, 33–45. [Google Scholar] [CrossRef]
- Leung, A.W.; Halestrap, A.P. Recent progress in elucidating the molecular mechanism of the mitochondrial permeability transition pore. Biochim. Biophys. Acta 2008, 1777, 946–952. [Google Scholar]
- Leung, A.W.; Varanyuwatana, P.; Halestrap, A.P. The mitochondrial phosphate carrier interacts with cyclophilin D and may play a key role in the permeability transition. J. Biol. Chem. 2008, 283, 26312–26323. [Google Scholar] [CrossRef]
- Dubourg, B.; Kamphausen, T.; Weiwad, M.; Jahreis, G.; Feunteun, J.; Fischer, G.; Modjtahedi, N. The human nuclear srcyp is a cell cycle-regulated cyclophilin. J. Biol. Chem. 2004, 279, 22322–22330. [Google Scholar]
- Teigelkamp, S.; Achsel, T.; Mundt, C.; Gothel, S.F.; Cronshagen, U.; Lane, W.S.; Marahiel, M.; Luhrmann, R. The 20kD protein of human [U4/U6.U5] tri-snRNPs is a novel cyclophilin that forms a complex with the U4/U6-specific 60kD and 90kD proteins. RNA 1998, 4, 127–141. [Google Scholar]
- Anderson, S.K.; Gallinger, S.; Roder, J.; Frey, J.; Young, H.A.; Ortaldo, J.R. A cyclophilin-related protein involved in the function of natural killer cells. Proc. Natl. Acad. Sci. USA 1993, 90, 542–546. [Google Scholar]
- Zhou, D.; Mei, Q.; Li, J.; He, H. Cyclophilin A and viral infections. Biochem. Biophys. Res. Commun. 2012, 424, 647–650. [Google Scholar] [CrossRef]
- Satoh, K.; Shimokawa, H.; Berk, B.C. Cyclophilin A: Promising new target in cardiovascular therapy. Circ. J. 2010, 74, 2249–2256. [Google Scholar] [CrossRef]
- Obchoei, S.; Wongkhan, S.; Wongkham, C.; Li, M.; Yao, Q.; Chen, C. Cyclophilin A: Potential functions and therapeutic target for human cancer. Med. Sci. Monit. 2009, 15, RA221–RA232. [Google Scholar]
- Luban, J.; Bossolt, K.L.; Franke, E.K.; Kalpana, G.V.; Goff, S.P. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 1993, 73, 1067–1078. [Google Scholar] [CrossRef]
- Watashi, K.; Ishii, N.; Hijikata, M.; Inoue, D.; Murata, T.; Miyanari, Y.; Shimotohno, K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 2005, 19, 111–122. [Google Scholar] [CrossRef]
- Bose, S.; Mathur, M.; Bates, P.; Joshi, N.; Banerjee, A.K. Requirement for cyclophilin a for the replication of vesicular stomatitis virus new jersey serotype. J. Gen. Virol. 2003, 84, 1687–1699. [Google Scholar] [CrossRef]
- Bienkowska-Haba, M.; Patel, H.D.; Sapp, M. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog. 2009, 5, e1000524. [Google Scholar] [CrossRef]
- Castro, A.P.; Carvalho, T.M.; Moussatche, N.; Damaso, C.R. Redistribution of cyclophilin a to viral factories during vaccinia virus infection and its incorporation into mature particles. J. Virol. 2003, 77, 9052–9068. [Google Scholar] [CrossRef]
- Billich, A.; Hammerschmid, F.; Peichl, P.; Wenger, R.; Zenke, G.; Quesniaux, V.; Rosenwirth, B. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus (HIV) type 1: Interference with hiv protein-cyclophilin a interactions. J. Virol. 1995, 69, 2451–2461. [Google Scholar]
- Ptak, R.G.; Gallay, P.A.; Jochmans, D.; Halestrap, A.P.; Ruegg, U.T.; Pallansch, L.A.; Bobardt, M.D.; de Bethune, M.P.; Neyts, J.; de Clercq, E.; et al. Inhibition of human immunodeficiency virus type 1 replication in human cells by Debio-025, a novel cyclophilin binding agent. Antimicrob. Agents Chemother. 2008, 52, 1302–1317. [Google Scholar] [CrossRef]
- Goto, K.; Watashi, K.; Murata, T.; Hishiki, T.; Hijikata, M.; Shimotohno, K. Evaluation of the anti-hepatitis C virus effects of cyclophilin inhibitors, cyclosporin A, and NIM811. Biochem. Biophys. Res. Commun. 2006, 343, 879–884. [Google Scholar] [CrossRef]
- Hopkins, S.; Scorneaux, B.; Huang, Z.; Murray, M.G.; Wring, S.; Smitley, C.; Harris, R.; Erdmann, F.; Fischer, G.; Ribeill, Y. Scy-635, a novel nonimmunosuppressive analog of cyclosporine that exhibits potent inhibition of hepatitis C virus RNA replication in vitro. Antimicrob. Agents Chemother. 2010, 54, 660–672. [Google Scholar] [CrossRef]
- Paeshuyse, J.; Kaul, A.; de Clercq, E.; Rosenwirth, B.; Dumont, J.M.; Scalfaro, P.; Bartenschlager, R.; Neyts, J. The non-immunosuppressive cyclosporin Debio-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology 2006, 43, 761–770. [Google Scholar] [CrossRef]
- de Wilde, A.H.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Thiel, V.; Narayanan, K.; Makino, S.; Snijder, E.J.; van Hemert, M.J. Cyclosporin a inhibits the replication of diverse coronaviruses. J. Gen. Virol. 2011, 92, 2542–2548. [Google Scholar] [CrossRef]
- Pfefferle, S.; Schopf, J.; Kogl, M.; Friedel, C.C.; Muller, M.A.; Carbajo-Lozoya, J.; Stellberger, T.; von Dall’Armi, E.; Herzog, P.; Kallies, S.; et al. The SARS-coronavirus-host interactome: Identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011, 7, e1002331. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Sato, Y.; Osawa, S.; Inoue, M.; Tanaka, S.; Sasaki, T. Suppression of feline coronavirus replication in vitro by cyclosporin A. Vet. Res. 2012, 43, e41. [Google Scholar] [CrossRef]
- Pedersen, N.C. A review of feline infectious peritonitis virus infection: 1963–2008. J. Feline Med. Surg. 2009, 11, 225–258. [Google Scholar] [CrossRef]
- Pedersen, N.C.; Boyle, J.F.; Floyd, K.; Fudge, A.; Barker, J. An enteric coronavirus infection of cats and its relationship to feline infectious peritonitis. Am. J. Vet. Res. 1981, 42, 368–377. [Google Scholar]
- Rottier, P.J.; Nakamura, K.; Schellen, P.; Volders, H.; Haijema, B.J. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J. Virol. 2005, 79, 14122–14130. [Google Scholar] [CrossRef]
- Hartmann, K.; Ritz, S. Treatment of cats with feline infectious peritonitis. Vet. Immunol. Immunopathol. 2008, 123, 172–175. [Google Scholar] [CrossRef]
- Tanaka, Y.; Osawa, S.; Inoue, M.; Tanaka, S.; Sato, Y. Inhibition of Feline Infectious Peritonitis Viral Replication by Cyclosporine. In Proceedings of the 29th Annual Meeting of the American Society for Virology, Bozeman, MN, USA, 17–21 July 2010. Abstract 55.
- de Wilde, A.H.; Li, Y.; van der Meer, Y.; Vuagniaux, G.; Lysek, R.; Fang, Y.; Snijder, E.J.; van Hemert, M.J. Cyclophilin inhibitors block arterivirus replication by interfering with viral RNA synthesis. J. Virol. 2013, 87, 1454–1464. [Google Scholar] [CrossRef]
- Kambara, H.; Tani, H.; Mori, Y.; Abe, T.; Katoh, H.; Fukuhara, T.; Taguwa, S.; Moriishi, K.; Matsuura, Y. Involvement of cyclophilin B in the replication of Japanese encephalitis virus. Virology 2011, 412, 211–219. [Google Scholar] [CrossRef]
- Qing, M.; Yang, F.; Zhang, B.; Zou, G.; Robida, J.M.; Yuan, Z.; Tang, H.; Shi, P.Y. Cyclosporine inhibits flavivirus replication through blocking the interaction between host cyclophilins and viral NS5 protein. Antimicrob. Agents Chemother. 2009, 53, 3226–3235. [Google Scholar] [CrossRef]
- Wathelet, M.G.; Orr, M.; Frieman, M.B.; Baric, R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: Role of nsp1 and rational design of an attenuated strain. J. Virol. 2007, 81, 11620–11633. [Google Scholar] [CrossRef]
- Zust, R.; Cervantes-Barragan, L.; Kuri, T.; Blakqori, G.; Weber, F.; Ludewig, B.; Thiel, V. Coronavirus non-structural protein 1 is a major pathogenicity factor: Implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007, 3, e109. [Google Scholar] [CrossRef]
- Brockway, S.M.; Denison, M.R. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology 2005, 340, 209–223. [Google Scholar] [CrossRef]
- Nagy, P.D.; Wang, R.Y.; Pogany, J.; Hafren, A.; Makinen, K. Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology 2011, 411, 374–382. [Google Scholar] [CrossRef]
- Coelmont, L.; Hanoulle, X.; Chatterji, U.; Berger, C.; Snoeck, J.; Bobardt, M.; Lim, P.; Vliegen, I.; Paeshuyse, J.; Vuagniaux, G.; et al. DEB025 (Alisporivir) inhibits hepatitis C virus replication by preventing a cyclophilin a induced cis-trans isomerisation in domain II of NS5A. PLoS One 2010, 5, e13687. [Google Scholar]
- Kaul, A.; Stauffer, S.; Berger, C.; Pertel, T.; Schmitt, J.; Kallis, S.; Zayas, M.; Lohmann, V.; Luban, J.; Bartenschlager, R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009, 5, e1000546. [Google Scholar] [CrossRef]
- Yang, F.; Robotham, J.M.; Grise, H.; Frausto, S.; Madan, V.; Zayas, M.; Bartenschlager, R.; Robinson, M.; Greenstein, A.E.; Nag, A.; et al. A major determinant of cyclophilin dependence and cyclosporine susceptibility of hepatitis C virus identified by a genetic approach. PLoS Pathog. 2010, 6, e1001118. [Google Scholar] [CrossRef]
- Yurchenko, V.; Zybarth, G.; O’Connor, M.; Dai, W.W.; Franchin, G.; Hao, T.; Guo, H.; Hung, H.C.; Toole, B.; Gallay, P.; et al. Active site residues of cyclophilin A are crucial for its signaling activity via CD147. J. Biol. Chem. 2002, 277, 22959–22965. [Google Scholar] [CrossRef]
- Chen, Z.; Mi, L.; Xu, J.; Yu, J.; Wang, X.; Jiang, J.; Xing, J.; Shang, P.; Qian, A.; Li, Y.; et al. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2005, 191, 755–760. [Google Scholar] [CrossRef]
- Gaither, L.A.; Borawski, J.; Anderson, L.J.; Balabanis, K.A.; Devay, P.; Joberty, G.; Rau, C.; Schirle, M.; Bouwmeester, T.; Mickanin, C.; et al. Multiple cyclophilins involved in different cellular pathways mediate HCV replication. Virology 2010, 397, 43–55. [Google Scholar]
- Pushkarsky, T.; Zybarth, G.; Dubrovsky, L.; Yurchenko, V.; Tang, H.; Guo, H.; Toole, B.; Sherry, B.; Bukrinsky, M. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. USA 2001, 98, 6360–6365. [Google Scholar]
- An, P.; Wang, L.H.; Hutcheson-Dilks, H.; Nelson, G.; Donfield, S.; Goedert, J.J.; Rinaldo, C.R.; Buchbinder, S.; Kirk, G.D.; O’Brien, S.J.; et al. Regulatory polymorphisms in the cyclophilin A gene, PPIA, accelerate progression to AIDS. PLoS Pathog. 2007, 3, e88. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, H. Contribution of cyclophilin A to determination of simian immunodeficiency virus tropism: A progress update. Vaccine 2010, 28 (Suppl. 2), B51–B54. [Google Scholar] [CrossRef]
- Provencher, V.M.; Coccaro, E.; Lacasse, J.J.; Schang, L.M. Antiviral drugs that target cellular proteins may play major roles in combating hiv resistance. Curr. Pharm. Des. 2004, 10, 4081–4101. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tanaka, Y.; Sato, Y.; Sasaki, T. Suppression of Coronavirus Replication by Cyclophilin Inhibitors. Viruses 2013, 5, 1250-1260. https://doi.org/10.3390/v5051250
Tanaka Y, Sato Y, Sasaki T. Suppression of Coronavirus Replication by Cyclophilin Inhibitors. Viruses. 2013; 5(5):1250-1260. https://doi.org/10.3390/v5051250
Chicago/Turabian StyleTanaka, Yoshikazu, Yuka Sato, and Takashi Sasaki. 2013. "Suppression of Coronavirus Replication by Cyclophilin Inhibitors" Viruses 5, no. 5: 1250-1260. https://doi.org/10.3390/v5051250




