Deletions within the 3' Non-Translated Region of Alfalfa mosaic virus RNA4 Do Not Affect Replication but Significantly Reduce Long-Distance Movement of Chimeric Tobacco mosaic virus
Abstract
:1. Introduction
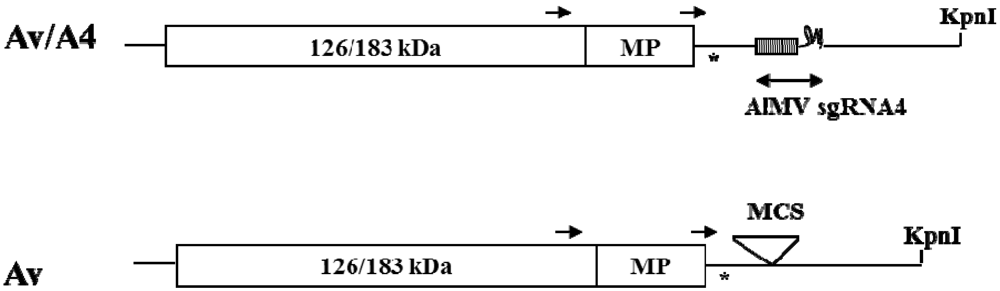
2. Results and Discussion
2.1. Effect of Deletions within the 3′ NTR of AlMV sgRNA4 on Chimeric Virus Replication and CP Accumulation

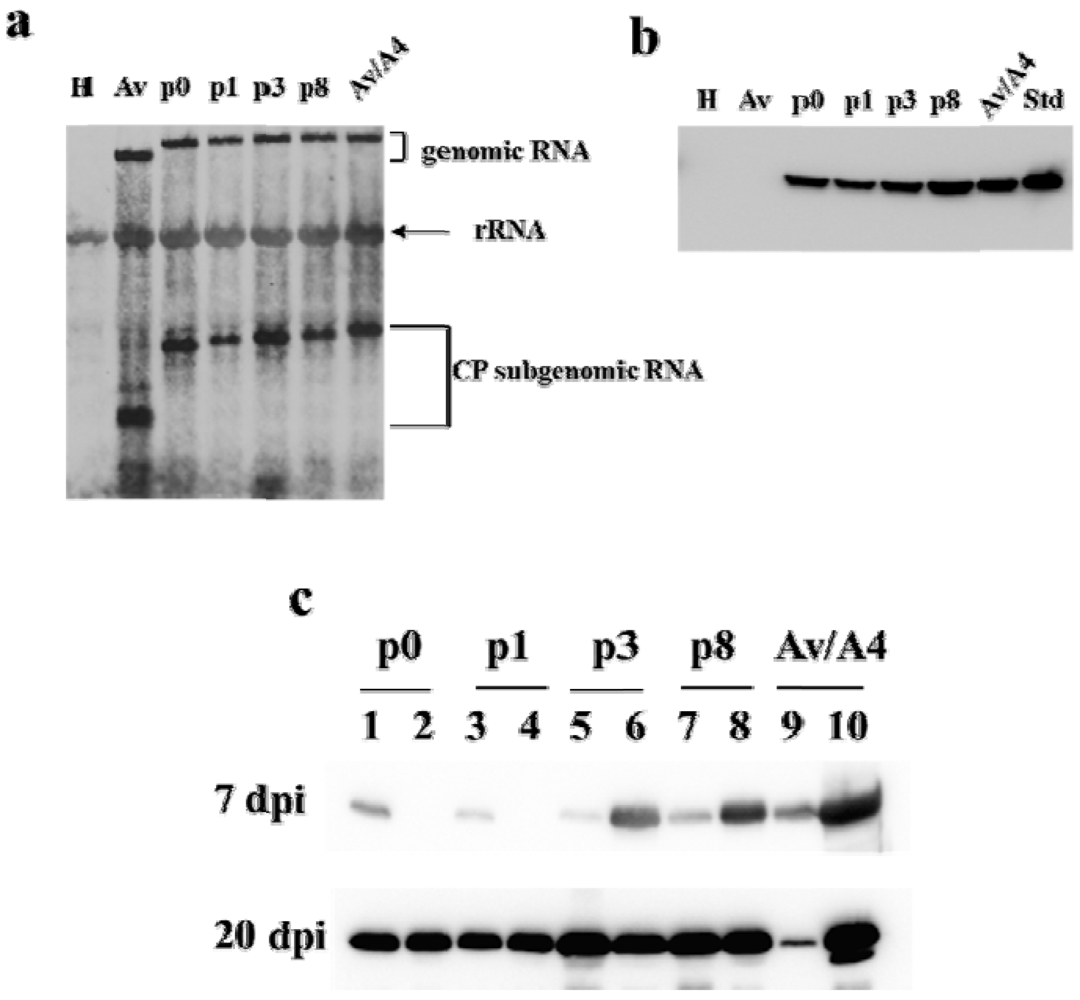
2.2. Symptom Expression, Encapsidation and Long-Distance Movement by AlMV sgRNA4 Mutants
2.3. Characterization of Purified Mutant Virus Particles
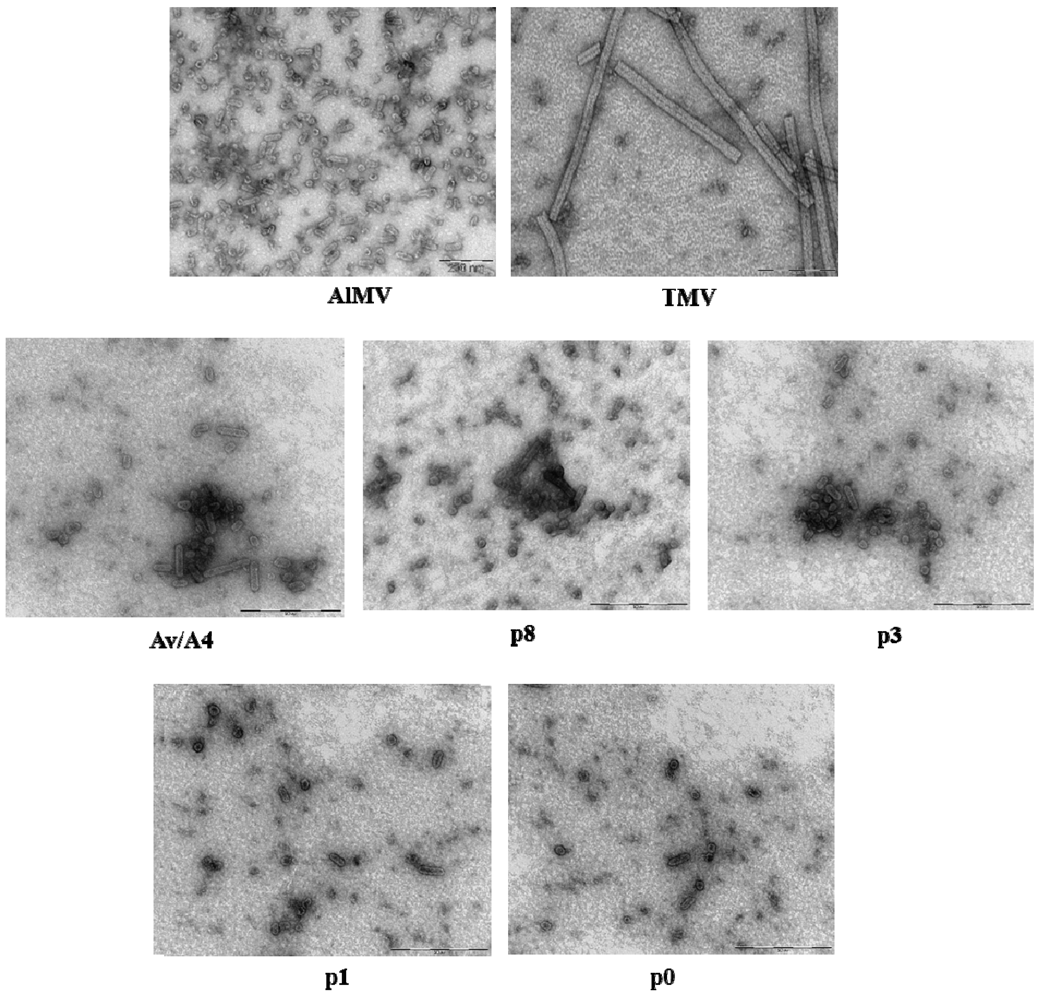
| Construct | Particle size (nm) a | |||||
|---|---|---|---|---|---|---|
| 95 nm | 70 | 62 | 44–48 | 30–35 | 16–24 | |
| Av/A4 | + | + | + | + | + | + |
| P8 | + | + | − | + | + | + |
| P3 | − | − | + | + | + | + |
| P1 | − | − | − | + | + | + |
| P0 | − | − | − | + | + | + |
3. Experimental Section
3.1. DNA Constructs
3.2. In Vitro Transcription and Protoplast Inoculation
3.3. Plant Inoculation and Virus Purification
3.4. Northern Blot Hybridization
3.5. Protein Analysis
3.6. Examination of Virion Stability in Plant Sap
3.7. Transmission Electron Microscopy
4. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Turner, D.R.; Joyce, L.E.; Butler, P.J. The tobacco mosaic virus assembly origin RNA. Functional characteristics defined by directed mutagenesis. J. Mol. Biol. 1988, 203, 531–547. [Google Scholar] [CrossRef]
- Zimmern, D. The nucleotide sequence at the origin for assembly on tobacco mosaic virus RNA. Cell 1977, 11, 463–482. [Google Scholar] [CrossRef]
- Choi, Y.G.; Rao, A.L. Molecular studies on bromovirus capsid protein. VII. Selective packaging on BMV RNA4 by specific N-terminal arginine residues. Virology 2000, 275, 207–217. [Google Scholar]
- Damayanti, T.A.; Tsukaguchi, S.; Mise, K.; Okuno, T. cis-acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts. J. Virol. 2003, 77, 9979–9986. [Google Scholar] [CrossRef]
- Gowda, S.; Satyanarayana, T.; Ayllon, M.A.; Moreno, P.; Flores, R.; Dawson, W.O. The conserved structures of the 5′ nontranslated region of Citrus tristeza virus are involved in replication and virion assembly. Virology 2003, 317, 50–64. [Google Scholar] [CrossRef]
- Qu, F.; Morris, T.J. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J. Virol. 1997, 71, 1428–1435. [Google Scholar]
- Hemmer, O.; Dunoyer, P.; Richards, K.; Fritsch, C. Mapping of viral RNA sequences required for assembly of peanut clump virus particles. J. Gen. Virol. 2003, 84, 2585–2594. [Google Scholar] [CrossRef]
- Thole, V.; Miglino, R.; Bol, J.F. Amino acids of alfalfa mosaic virus coat protein that direct formation of unusually long virus particles. J. Gen. Virol. 1998, 79, 3139–3143. [Google Scholar]
- De Graaff, M.; Man in't Veld, M.R.; Jaspars, E.M. In vitro evidence that the coat protein of alfalfa mosaic virus plays a direct role in the regulation of plus and minus RNA synthesis: Implications for the life cycle of alfalfa mosaic virus. Virology 1995, 208, 583–589. [Google Scholar] [CrossRef]
- Van der Kuyl, AC.; Neeleman, L.; Bol, J.F. Role of alfalfa mosaic virus coat protein in regulation of the balance between viral plus and minus strand RNA synthesis. Virology 1991, 185, 496–499. [Google Scholar] [CrossRef]
- Sanchez-Navarro, J.A.; Bol, J.F. Role of the alfalfa mosaic virus movement protein and coat protein in virus transport. Mol. Plant Microbe Interact. 2001, 14, 1051–1062. [Google Scholar] [CrossRef]
- Stussi-Garaud, C.; Garaud, G.J.; Berna, A.; Godefroy-Colburn, T. In situ location of an alfalfa mosaic virus nonstructural protein in plant cell walls: Correlation with virus transport. J. Gen. Virol. 1987, 68, 1179–1184. [Google Scholar]
- Bol, J.F.; van Vloten-Doting, L.; Jaspars, E.M. A functional equivalence of top component a RNA and coat protein in the initiation of infection by alfalfa mosaic virus. Virology 1971, 46, 73–85. [Google Scholar]
- van der Vossen, E.A.; Neeleman, L.; Bol, J.F. Early and late functions of alfalfa mosaic virus coat protein can be mutated separately. Virology 1994, 202, 891–903. [Google Scholar] [CrossRef]
- Jaspars, E.M. Plant viruses with a multipartite genome. Adv. Virus Res. 1974, 19, 37–149. [Google Scholar] [CrossRef]
- Loesch-Fries, L.S.; Jarvis, N.P.; Krahn, K.J.; Nelson, S.E.; Hall, T.C. Expression of alfalfa mosaic virus RNA 4 cDNA transcripts in vitro and in vivo. Virology 1985, 146, 177–187. [Google Scholar] [CrossRef]
- Neeleman, L.; Van der Vossen, E.A.; Bol, J.F. Infection of tobacco with alfalfa mosaic virus cDNAs sheds light on the early function of the coat protein. Virology 1993, 196, 883–887. [Google Scholar] [CrossRef]
- Neeleman, L.; Bol, J.F. Cis-acting functions of alfalfa mosaic virus proteins involved in replication and encapsidation of viral RNA. Virology 1999, 254, 324–333. [Google Scholar] [CrossRef]
- Vlot, A.C.; Neeleman, L.; Linthorst, H.J.; Bol, J.F. Role of the 3′-untranslated regions of alfalfa mosaic virus RNAs in the formation of a transiently expressed replicase in plants and in the assembly of virions. J. Virol. 2001, 75, 6440–6449. [Google Scholar] [CrossRef]
- Spitsin, S.; Steplewski, K.; Fleysh, N.; Belanger, H.; Mikheeva, T.; Shivprasad, S.; Dawson, W.; Koprowski, H.; Yusibov, V. Expression of alfalfa mosaic virus coat protein in tobacco mosaic virus (TMV) deficient in the production of its native coat protein supports long-distance movement of a chimeric TMV. Proc. Natl. Acad. Sci. USA 1999, 96, 2549–2553. [Google Scholar] [CrossRef]
- Neeleman, L.; Linthorst, H.J.; Bol, J.F. Efficient translation of alfamovirus RNAs requires the binding of coat protein dimers to the 3′ termini of the viral RNAs. J. Gen. Virol. 2004, 85, 231–240. [Google Scholar] [CrossRef]
- Yusibov, V.; Modelska, A.; Steplewski, K.; Agadjanyan, M.; Weiner, D.; Hooper, D.C.; Koprowski, H. Antigens produced in plants by infection with chimeric plant viruses immunize against rabies virus and HIV-1. Proc. Natl. Acad. Sci. USA 1997, 94, 5784–5788. [Google Scholar] [CrossRef]
- Choi, Y.G.; Dreher, T.W.; Rao, A.L. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. USA 2002, 99, 655–660. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Lewandowski, D.J.; Dawson, W.O. Deletion of internal sequences results in tobacco mosaic virus defective RNAs that accumulate to high levels without interfering with replication of the helper virus. Virology 1998, 251, 427–437. [Google Scholar] [CrossRef]
- Fitchen, J.; Beachy, R.N.; Hein, M.B. Plant virus expressing hybrid coat protein with added murine epitope elicits autoantibody response. Vaccine 1995, 13, 1051–1057. [Google Scholar] [CrossRef]
- Gooding, G.V., Jr.; Hebert, T.T. A simple technique for purification of tobacco mosaic virus in large quantities. Phytopathology 1967, 57, 1285. [Google Scholar]
- Annamalai, P.; Rao, A.L. In vivo packaging of brome mosaic virus RNA3, but not RNAs 1 and 2, is dependent on a cis-acting 3′ tRNA-like structure. J. Virol. 2007, 81, 173–181. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Roy, G.; Fedorkin, O.; Fujiki, M.; Skarjinskaia, M.; Knapp, E.; Rabindran, S.; Yusibov, V. Deletions within the 3' Non-Translated Region of Alfalfa mosaic virus RNA4 Do Not Affect Replication but Significantly Reduce Long-Distance Movement of Chimeric Tobacco mosaic virus. Viruses 2013, 5, 1802-1814. https://doi.org/10.3390/v5071802
Roy G, Fedorkin O, Fujiki M, Skarjinskaia M, Knapp E, Rabindran S, Yusibov V. Deletions within the 3' Non-Translated Region of Alfalfa mosaic virus RNA4 Do Not Affect Replication but Significantly Reduce Long-Distance Movement of Chimeric Tobacco mosaic virus. Viruses. 2013; 5(7):1802-1814. https://doi.org/10.3390/v5071802
Chicago/Turabian StyleRoy, Gourgopal, Oleg Fedorkin, Masaaki Fujiki, Marina Skarjinskaia, Elisabeth Knapp, Shailaja Rabindran, and Vidadi Yusibov. 2013. "Deletions within the 3' Non-Translated Region of Alfalfa mosaic virus RNA4 Do Not Affect Replication but Significantly Reduce Long-Distance Movement of Chimeric Tobacco mosaic virus" Viruses 5, no. 7: 1802-1814. https://doi.org/10.3390/v5071802




