Retroviral Infections in Sheep and Goats: Small Ruminant Lentiviruses and Host Interaction
Abstract
:1. Small Ruminant Lentiviruses (SRLVs)
1.1. SRLV Infection and Disease
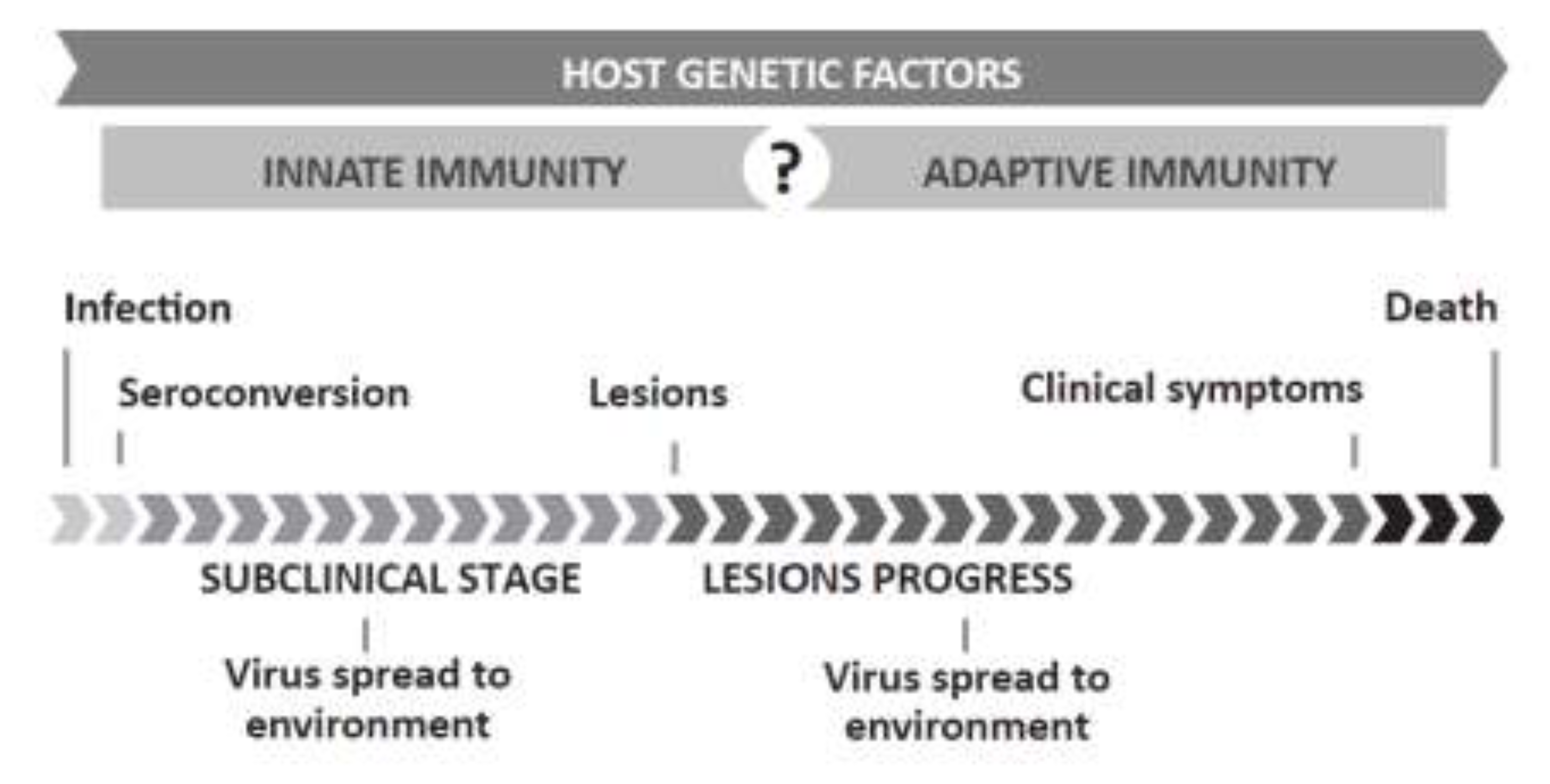
1.2. Relevance and Control of SRLVs
2. Host Genetic Resources
3. Host-Viral Pathogen Interaction
| HOST | GENOTYPING | ||||
|---|---|---|---|---|---|
| Population stratification | Often present in livestock due to breeding practices | Bias/Partiality | Case samples are treated with preference | ||
| Sample size | Affects power to detect association | Marker density | Often few markers are analyzed per gene | ||
| Phenotype | Description must be accurate | Marker frequency | Frequency of marker variants affect odds of detecting association | ||
| Age | Older animals have been exposed for a longer time | STATISTICAL ANALYSIS | |||
| Gene effect | Genes involved have small/moderate effects | Power | Depends on sample size, marker frequency and gene effect | ||
| Presence of other diseases | May facilitate SRLV pathogenesis | Gene interaction | Often unaccounted for | ||
| SRLVs | Confounding factors | Failure to account for them may lead to erroneous interpretations | |||
| Different virus strains | Different virulence and host/organ spectrum complicates research; | Multiple corrections | Necessary yet may lead to reject real associations | ||
| Strain variability may affect detection of infected individuals | Consistency/replicability | Results must be replicated in different populations/ | |||
| ENVIRONMENTAL FACTORS | Can results be replicated in a different population? | ||||
| Husbandry | Prolonged and crowded housing enhances infection | ||||
4. Evidence Pointing to the Existence of Host Genetics Controlling SRLV Pathogenesis
4.1. Infection
4.2. Clinical Disease
5. Host Genetic Factors Involved in SRLV-Induced Pathogenesis
5.1. Major Histocompatibility Complex (MHC)
| Region | Variant | Typing method/marker | Species/Breed(s) studied | Reference |
|---|---|---|---|---|
| Class I | allele CLA Be7 | Alloantisera | Goat/Saanen | [97] |
| Allele OMHC1* 205 | Microsatellite | Sheep/Latxa | [96] | |
| Class II | Alleles DRB1*0403- and DRB1*07012 and various amino acid positions | Cloning and direct sequencing | Sheep/Rambouillet, Polipay & Columbia | [64] |
| DRB1*0325 | PCR-Sequence-Based Typing | Sheep/Latxa | [95] | |
| Allele DRB2* 275 | Microsatellite | Sheep/Latxa | [96] |
5.2. Antibody and T Cell Response
5.3. Cytokines and Receptors
5.4. Innate Immunity and Restriction Factors
| Gene symbol | Gene | Species | Methods | Analyzed material | Parameter analysed | References |
|---|---|---|---|---|---|---|
| IL1β | Interleukin-1beta | Sheep | Semiquantitative RT-PCR 1 | Lung | Clinical disease | [114] |
| IL2/IL2R | Interleukin-2/Interleukin-2 receptor | Sheep | Semiquantitative RT-PCR, qPCR 2 | Lung, PBMCs 3, lymph node leukocytes | Infection, Clinical disease | [114,118,120] |
| Goat | In situ hybridization | Joints | Infection, Clinical disease | [113] | ||
| IL4 | Interleukin-4 | Sheep | Semiquantitative RT-PCR | Lung | Clinical disease | [114] |
| Goat | Semiquantitative RT-PCR | PBMCs | Clinical disease | [20] | ||
| IL6 | Interleukin-6 | Sheep | RT-PCR | Alveolar macrophages | Infection, Clinical disease | [116] |
| IL8 | Interleukin-8 | Sheep | RT-qPCR, in situ hybridization, Northern blot, Semiquantitative RT-PCR | BALF, alveolar macrophages, | Infection, Clinical disease | [115,117,119] |
| Goat | In situ hybridization | Macrophages | Infection | [20] | ||
| IL10 | Interleukin-10 | Sheep | Semiquantitative RT-PCR | Lung, alveolar macrophages | Infection, Clinical disease | [114,116] |
| IFNy | Interferon-gamma | Sheep | Semiquantitative RT-PCR | Lung | Clinical disease | [114] |
| Goat | Semiquantitative RT-PCR, in situ hybridization | PBMCs, joints | Clinical disease | [19,113] | ||
| TNFα | Tumor Necrosis factor-alpha | Sheep | qPCR | Lung, udder | Clinical disease | [120] |
| TGF-β1 | Tumor growth factor beta-1 | Sheep | Semiquantitative RT-PCR | Alveolar macrophages | Infection, Clinical disease | [116] |
| Goat | In situ hybridization | Macrophages | Infection | [20] | ||
| MCP-1 | Monocyte chemoattractant protein 1 | Goat | In situ hybridization | Macrophages, joints | Infection ,Clinical disease | [20,113] |
| GM-CSF | Granulocyte macrophage stimulating factor | Sheep | Semiquantitative RT-PCR | Lung, alveolar macrophages | Infection, Clinical disease | [114,116] |
| Goat | In situ hybridization | Macrophages | Infection | [20] | ||
| CCR5 | Chemokine (C-C motif) Receptor 5 | Sheep | Cloning & sequencing, qPCR | PBMCs, lung | Infection, Clinical disease | [120,121] |
6. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Patel, J.R.; Heldens, J.G.M.; Bakonyi, T.; Rusvai, M. Important mammalian veterinary viral immunodiseases and their control. Vaccine 2012, 30, 1767–1781. [Google Scholar] [CrossRef]
- Leroux, C.; Cruz, J.C.M.; Mornex, J.F. SRLVs: A genetic continuum of lentiviral species in sheep and goats with cumulative evidence of cross species transmission. Curr. HIV Res. 2010, 8, 94–100. [Google Scholar]
- Leroux, C.; Vuillermoz, S.; Mornex, J.F.; Greenland, T. Genomic heterogeneity in the pol region of ovine lentiviruses obtained from bronchoalveolar cells of infected sheep from France. J. Gen. Virol. 1995, 76, 1533–1537. [Google Scholar] [CrossRef]
- Gorrell, M.D.; Brandon, M.R.; Sheffer, D.; Adams, R.J.; Narayan, O. Ovine lentivirus is macrophagetropic and does not replicate productively in T lymphocytes. J. Virol. 1992, 66, 2679–2688. [Google Scholar]
- Ryan, S.; Tiley, L.; McConnell, I.; Blacklaws, B. Infection of dendritic cells by the Maedi-Visna lentivirus. J. Virol. 2000, 74, 10096–10103. [Google Scholar] [CrossRef]
- Christodoulopoulos, G. Maedi-Visna: Clinical review and short reference on the disease status in Mediterranean countries. Small Rumin. Res. 2006, 62, 47–53. [Google Scholar] [CrossRef]
- Blacklaws, B.A. Small ruminant lentiviruses: Immunopathogenesis of Visna-Maedi and caprine arthritis and encephalitis virus. Comp. Immunol. Microbiol. Infect.Dis. 2012, 35, 259–269. [Google Scholar] [CrossRef]
- Rowe, J.D.; East, N.E. Risk factors for transmission and methods for control of caprine arthritis-encephalitis virus infection. Vet. Clin. North Am. Food Anim. Pract. 1997, 13, 35–53. [Google Scholar]
- Blacklaws, B.A.; Berriatúa, E.; Torsteinsdottir, S.; Watt, N.J.; De Andrés, D.; Klein, D.; Harkiss, G.D. Transmission of small ruminant lentiviruses. Vet. Microbiol. 2004, 101, 199–208. [Google Scholar] [CrossRef]
- Pépin, M.; Vitu, C.; Russo, P.; Mornex, J.F.; Peterhans, E. Maedi-Visna Virus infection in sheep: A review. Vet. Res. 1998, 29, 1–53. [Google Scholar]
- Peterhans, E.; Greenland, T.; Badiola, J.; Harkiss, G.; Bertoni, G.; Amorena, B.; Eliaszewicz, M.; Juste, R.A.; Krassnig, R.; Lafont, J.P.; et al. Routes of transmission and consequences of small ruminant lentiviruses (SRLVs) infection and eradication schemes. Vet. Res. 2004, 35, 257–274. [Google Scholar]
- Leginagoikoa, I.; Daltabuit-Test, M.; Alvarez, V.; Arranz, J.; Juste, R.A.; Amorena, B.; de Andrés, D.; Luján, L.L.; Badiola, J.J.; Berriatúa, E. Horizontal Maedi-Visna Virus (MVV) infection in adult dairy-sheep raised under varying MVV-infection pressures investigated by ELISA and PCR. Res. Vet. Sci. 2006, 80, 235–241. [Google Scholar] [CrossRef]
- Gufler, H.; Gasteiner, J.; Lombardo, D.; Stifter, E.; Krassnig, R. Serological study of small ruminant lentivirus in goats in Italy. Small Rumin. Res. 2007, 73, 169–173. [Google Scholar] [CrossRef]
- Herrmann-Hoesing, L.M.; Palmer, G.G.H.; Knowles, D.P.D. Evidence of proviral clearance following postpartum transmission of an ovine lentivirus. Virology 2007, 362, 226–234. [Google Scholar] [CrossRef]
- Broughton-neiswanger, L.E.; White, S.N.; Knowles, D.P.; Mousel, M.R.; Lewis, G.S.; Herndon, D.R.; Herrmann-Hoesing, L.M. Non-maternal transmission is the major mode of ovine lentivirus transmission in a ewe flock: A molecular epidemiology study. Infect. Genet. Evol. 2010, 10, 998–1007. [Google Scholar] [CrossRef]
- Álvarez, V.; Arranz, J.; Daltabuit-Test, M.; Leginagoikoa, I.; Juste, R.A.; Amorena, B.; de Andrés, D.; Luján, L.L.; Badiola, J.J.; Berriatúa, E. Relative contribution of colostrum from Maedi-Visna Virus (MVV) infected ewes to MVV-seroprevalence in lambs. Res. Vet. Sci. 2005, 78, 237–243. [Google Scholar] [CrossRef]
- Thormar, H. Maedi-Visna Virus and its relationship to human immunodeficiency virus. AIDS Rev. 2005, 7, 233–245. [Google Scholar]
- Leroux, C.; Mornex, J.F. Retroviral infections in sheep and the associated diseases. Small Rumin. Res. 2008, 76, 68–76. [Google Scholar] [CrossRef]
- Cheevers, W.P.; Beyer, J.C.; Knowles, D.P. Type 1 and type 2 cytokine gene expression by viral gp135 surface protein-activated T lymphocytes in caprine arthritis-encephalitis lentivirus infection. J. Virol. 1997, 71, 6259–6263. [Google Scholar]
- Lechner, F.; Machado, J.; Bertoni, G.; Seow, H.F.; Dobbelaere, D.A.E.; Peterhans, E. Caprine arthritis encephalitis virus dysregulates the expression of cytokines in macrophages. J. Virol. 1997, 71, 7488–7497. [Google Scholar]
- Ravazzolo, A.P.; Nenci, C.; Vogt, H.R.; Waldvogel, A.; Obexer-Ruff, G.; Peterhans, E.; Bertoni, G. Viral load, organ distribution, histopathological lesions, and cytokine mRNA expression in goats infected with a molecular clone of the caprine arthritis encephalitis virus. Virology 2006, 350, 116–127. [Google Scholar] [CrossRef]
- Benavides, J.; García-Pariente, C.; Fuertes, M.; Ferreras, M.C.; García-Marín, J.F.; Juste, R.A. Maedi-Visna: The meningoencephalitis in naturally occurring cases. J. Comp. Pathol. 2009, 141, 1–11. [Google Scholar] [CrossRef]
- Heaton, M.P.M.; Clawson, M.L.M.; Chitko-Mckown, C.G.; Leymaster, K.A.; Smith, T.P.L.; Harhay, G.P.; White, S.N.; Herrmann-Hoesing, L.M.; Mousel, M.R.; Lewis, G.S.; et al. Reduced lentivirus susceptibility in sheep with TMEM154 mutations. PLoS Genet. 2012, 8, e1002467. [Google Scholar] [CrossRef]
- Cheevers, W.P.; Ho, I.; Beyer, J.C. Immune response to caprine arthritis/encephalitis virus surface protein induced by coimmunization with recombinant vaccinia viruses expressing the caprine arthritis/encephalitis virus envelope gene and caprine interleukin-12. Vaccine 2000, 18, 2494–2503. [Google Scholar] [CrossRef]
- Torsteinsdóttir, S.; Carlsdóttir, H.M.; Svansson, V.; Matthíasdóttir, S.; Pétursson, G. Vaccination of sheep with Maedi-Visna Virus gag gene and protein, beneficial or harmful? Vaccine 2007, 25, 6713–6720. [Google Scholar] [CrossRef]
- Brodie, S.J.; Marcom, K.A.; Pearson, L.D.; Anderson, B.C.; de la Concha-Bermejillo, A.; Ellis, J.A.; DeMartini, J.C. Effects of virus load in the pathogenesis of lentivirus-induced lymphoid interstitial pneumonia. J. Infect. Dis. 1992, 166, 531–541. [Google Scholar] [CrossRef]
- Pétursson, G.; Matthíasdóttir, S.; Svansson, V.; Andrésdóttir, V.; Georgsson, G.; Martin, A.H.; Agnarsdóttir, G.; Gísladóttir, E.; Arnadóttir, S.; Högnadóttir, S.; et al. Mucosal vaccination with an attenuated Maedi-Visna Virus clone. Vaccine 2005, 23, 3223–3228. [Google Scholar] [CrossRef]
- Cheevers, W.P.; Snekvik, K.R.; Trujillo, J.D.; Kumpula-McWhirter, N.M.; Pretty On Top, K.J.; Knowles, D.P. Prime-boost vaccination with plasmid DNA encoding caprine-arthritis encephalitis lentivirus env and viral SU suppresses challenge virus and development of arthritis. Virology 2003, 306, 116–125. [Google Scholar] [CrossRef]
- Nenci, C.; Zahno, M.L.; Vogt, H.R.; Obexer-Ruff, G.; Doherr, M.G.; Zanoni, R.; Bertoni, G. Vaccination with a T-cell-priming Gag peptide of caprine arthritis encephalitis virus enhances virus replication transiently in vivo. J. Gen. Virol. 2007, 88, 1589–1593. [Google Scholar] [CrossRef]
- Blacklaws, B.; Bird, P.; McConnell, I. Early events in infection of lymphoid tissue by a lentivirus, Maedi-Visna. Trends Microbiol. 1995, 3, 434–440. [Google Scholar] [CrossRef]
- Berriatúa, E.; Álvarez, V.; Extramiana, B.; González, L.; Daltabuit, M.; Juste, R. Transmission and control implications of seroconversion to Maedi-Visna Virus in Basque dairy-sheep flocks. Prev. Vet. Med. 2003, 60, 265–279. [Google Scholar] [CrossRef]
- DeMartini, J.; de la Concha-Bermejillo, A.; Carlson, J.; Bowen, R. Diseases caused by Maedi-Visna and other ovine lentiviruses. In Breeding for Disease Resistance in Farm Animals; Axford, R.F.E., Bishop, S.C., Nicholas, F.W., Owen, J.B., Eds.; CAB International: Oxon, UK, 2000; pp. 301–324. [Google Scholar]
- Bishop, S.; Morris, C. Genetics of disease resistance in sheep and goats. Small Rumin. Res. 2007, 70, 48–59. [Google Scholar] [CrossRef]
- Torsteinsdottir, S.; Andresdottir, V.; Arnarson, H.; Petursson, G. Immune response to Maedi-Visna Virus. Front Biosci. 2007, 12, 1532–1543. [Google Scholar] [CrossRef]
- Lawson-Handley, L.; Byrne, K.; Santucci, F.; Townsend, S.; Taylor, M.; Bruford, M.W.; Hewitt, G.M. Genetic structure of European sheep breeds. Heredity 2007, 99, 620–631. [Google Scholar] [CrossRef]
- Kijas, J.W.; Townley, D.; Dalrymple, B.P.; Heaton, M.P.; Maddox, J.F.; Wilson, P.; Ingersoll, R.G.; Mcculloch, R.; Mcwilliam, S.; Tang, D.; et al. A Genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS One 2009, 4, e4668. [Google Scholar] [CrossRef]
- The International Sheep Genomics Consortium; Archibald, A.; Cockett, N.; Dalrymple, B.P.; Faraut, T.; Kijas, J.W.; Maddox, J.F.; Mcewan, J.C.; Oddy, V.H.; Raadsma, H.W.; et al. The sheep genome reference sequence: A work in progress. Anim. Genet. 2010, 2009, 449–453. [Google Scholar]
- Ibeagha-Awemu, E.M.; Kgwatalala, E.M.I.P.; Ibeagha, A.E.; Zhao, X. A critical analysis of disease-associated DNA polymorphisms in the genes of cattle, goat, sheep, and pig. Mamm. Genome 2008, 19, 226–245. [Google Scholar] [CrossRef]
- Becker, D.; Tetens, J.; Brunner, A.; Bürstel, D.; Ganter, M.; Kijas, J.; International Sheep Genomics Consortium; Drögemüller, C. Microphthalmia in Texel sheep is associated with a missense mutation in the paired-like homeodomain 3 (PITX3) gene. PLoS One 2010, 5, e8689. [Google Scholar] [CrossRef] [Green Version]
- Purdie, A.C.; Plain, K.M.; Begg, D.J.; de Silva, K.; Whittington, R.J.; de Silva, K. Candidate gene and genome-wide association studies of Mycobacterium avium subsp. paratuberculosis infection in cattle and sheep: A review. Comp. Immunol. Microbiol. Infect. Dis. 2011, 34, 197–208. [Google Scholar] [CrossRef]
- Silverman, E.K.; Palmer, L.J. Case-control association studies for the genetics of complex respiratory diseases. Am. J. Respir. Cell Mol. Biol. 2000, 22, 645–648. [Google Scholar] [CrossRef]
- Daly, A.K.; Day, C.P. Candidate gene case-control association studies: Advantages and potential pitfalls. Br. J. Clin. Pharmacol. 2001, 52, 489–499. [Google Scholar] [CrossRef]
- Campbell, H.; Rudan, I. Interpretation of genetic association studies in complex disease. Pharmacogenomics J. 2002, 2, 349–360. [Google Scholar] [CrossRef]
- Hirschhorn, J.N.; Lohmueller, K.; Byrne, E.; Hirschhorn, K. A comprehensive review of genetic association studies. Genet. Med. 2002, 4, 45–61. [Google Scholar] [CrossRef]
- Cardon, L.R.; Palmer, L.J. Population stratification and spurious allelic association. Lancet 2003, 361, 598–604. [Google Scholar] [CrossRef]
- Colhoun, H.M.; McKeigue, P.M.; Smith, G.D. Problems of reporting genetic associations with complex outcomes. Lancet 2003, 361, 865–872. [Google Scholar] [CrossRef]
- Cordell, H.J.; Clayton, D.G. Genetic association studies. Lancet 2005, 366, 1121–1131. [Google Scholar]
- Balding, D.J. A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 2006, 7, 781–791. [Google Scholar] [CrossRef]
- Narum, S.R. Beyond bonferroni: Less conservative analyses for conservation genetics. Conserv. Genet. 2006, 7, 783–787. [Google Scholar] [CrossRef]
- Bochud, P.Y.; Bochud, M.; Telenti, A.; Calandra, T. Innate immunogenetics: A tool for exploring new frontiers of host defence. Lancet Infect. Dis. 2007, 7, 531–542. [Google Scholar] [CrossRef]
- Abbas, A.K.; Lichtman, A.H. Basic Immunology, Updated Edition; Saunders Elsevier: Philadelphia, PA, USA, 2006. [Google Scholar]
- Maclachlan, N.; Dubovi, E.J. Fenner’s Veterinary Virology, 4th ed.; Elsevier: London, UK, 2011. [Google Scholar]
- Miyazawa, M.; Tsuji-kawahara, S.; Kanari, Y. Host genetic factors that control immune responses to retrovirus infections. Vaccine 2010, 26, 2981–2996. [Google Scholar]
- Kaur, G.; Mehra, N. Genetic determinants of HIV-1 infection and progression to AIDS: Immune response genes. Tissue Antigens 2009, 74, 373–385. [Google Scholar] [CrossRef]
- Brodie, S. Current concepts in the epizootiology, diagnosis, and economic importance of ovine progressive pneumonia in North America: A review. Small Rumin. Res. 1998, 27, 1–17. [Google Scholar] [CrossRef]
- Blacklaws, B.A.; Harkiss, G.D. Small ruminant lentiviruses and human immunodeficiency virus: Cousins that take a long view. Curr. HIV Res. 2010, 8, 26–52. [Google Scholar] [CrossRef]
- Stump, D.S.; VandeWoude, S. Animal models for HIV AIDS: A comparative review. Comp. Med. 2007, 57, 33–43. [Google Scholar]
- Angelopoulou, K.; Brellou, G.D.; Vlemmas, I.; Greenland, T. A novel deletion in the LTR region of a Greek small ruminant lentivirus may be associated with low pathogenicity. Virus Res. 2006, 118, 178–184. [Google Scholar] [CrossRef]
- Oskarsson, T.; Hreggvidsdóttir, H.S.; Agnarsdóttir, G.; Matthíasdóttir, S.; Ogmundsdóttir, M.H.; Jónsson, S.R.; Georgsson, G.; Ingvarsson, S.; Andrésson, O.S.; Andrésdóttir, V. Duplicated sequence motif in the long terminal repeat of Maedi-Visna Virus extends cell tropism and is associated. J. Virol. 2007, 81, 4052–4057. [Google Scholar] [CrossRef]
- Leginagoikoa, I.; Minguijón, E.; Juste, R.A.; Barandika, J.; Amorena, B.; de Andrés, D.; Badiola, J.J.; Luján, L.; Berriatua, E.; de Andrés, D. Effects of housing on the incidence of Visna/Maedi virus infection in sheep flocks. Res. Vet. Sci. 2010, 88, 415–421. [Google Scholar] [CrossRef]
- González, L.; Juste, R.A.; Cuervo, L.A.; Idigoras, I.; Saez de Ocariz, C. Pathological and epidemiological aspects of the coexis- tence of Maedi-Visna and sheep pulmonary adenomatosis. Res. Vet. Sci. 1993, 54, 140–146. [Google Scholar] [CrossRef]
- Andrésdóttir, V.; Tang, X.; Agnarsdóttir, G.; Andrésson, O.S.; Georgsson, G.; Skraban, R.; Torsteinsdóttir, S.; Rafnar, B.; Benediktsdóttir, E.; Matthíasdóttir, S.; Arnadóttir, S.; et al. Biological and genetic differences between lung- and brain-derived isolates of Maedi-Visna Virus. Virus Genes 1998, 16, 281–293. [Google Scholar] [CrossRef]
- Reina, R.; Grego, E.; Bertolotti, L.; de Meneghi, D.; Rosati, S. Genome analysis of small-ruminant lentivirus genotype E: A caprine lentivirus with natural deletions of the dUTPase subunit, vpr-like accessory gene, and 70-base-pair repeat of the U3 region. J. Virol. 2009, 83, 1152–1155. [Google Scholar] [CrossRef]
- Herrmann-Hoesing, L.M.; White, S.N.; Mousel, M.R.; Lewis, G.S.; Knowles, D.P. Ovine progressive pneumonia provirus levels associate with breed and Ovar-DRB1. Immunogenetics 2008, 60, 749–758. [Google Scholar] [CrossRef]
- Palsson, P. Maedi and visna in sheep. In Slow Virus Diseases of Animals and Man; Kimberlin, R.H., Ed.; North-Holland: Amsterdam, The Netherland, 1976; pp. 17–43. [Google Scholar]
- Straub, O.C. Maedi-Visna Virus infection in sheep. History and present knowledge. Comp. Immunol. Microbiol. Infect. Dis. 2004, 27, 1–5. [Google Scholar] [CrossRef]
- Cutlip, R.C.; Lehmkuhl, H.D.; Brogden, K.A.; Sacks, J.M. Breed susceptibility to ovine progressive pneumonia (Maedi/Visna) virus. Vet. Microbiol. 1986, 12, 283–288. [Google Scholar] [CrossRef]
- Houwers, D.J.; VIsscher, A.H.; Defise, P.R. Importance of ewe/lamb relationship and breed in the epidemiology of Maedi-Visna VIRUS infections. Res. Vet. Sci. 1989, 46, 5–8. [Google Scholar]
- Zink, M.C.; Johnson, L.K. Pathobiology of lentivirus infections of sheep and goats. Virus Res. 1994, 32, 139–154. [Google Scholar] [CrossRef]
- De la Concha-Bermejillo, A.; Brodie, S.J.; Magnus-Corral, S.; Bowen, R.A.; DeMartini, J.C. Pathologic and serologic responses of isogeneic twin lambs to phenotypically disinct lentiviruses. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1995, 8, 116–123. [Google Scholar]
- Gates, N.L.; Winward, L.D.; Gorham, J.R.; Shen, D.T. Serologic survey of prevalence of ovine progressive pneumonia in Idaho range sheep. J. Am. Vet. Med. Assoc. 1978, 173, 1575–1577. [Google Scholar]
- Keen, J.E.; Hungerford, L.L.; Wittum, T.E.; Kwang, J.; Littledike, E.T. Risk factors for seroprevalenceof ovine lentivirus in breeding ewe flocks in Nebraska, USA. Prev. Vet. Med. 1997, 30, 81–94. [Google Scholar] [CrossRef]
- Rowe, J.D.; East, N.E.; Franti, C.E.; Thurmond, M.C.; Pedersen, N.C.; Theilen, G.H. Risk factors associated with the incidence of seroconversion to caprine arthritis-encephalitis virus in goats on California dairies. Am. J. Vet. Res. 1992, 53, 2396–2403. [Google Scholar]
- Perk, K. Characteristics of ovine and caprine lentivirus infections. Leukemia 1995, 9, 98–100. [Google Scholar]
- Zhang, Z.; Watt, N.J.; Hopkins, J.; Harkiss, G.; Woodall, C.J. Quantitative analysis of Maedi-Visna Virus DNA load in peripheral blood monocytes and alveolar macrophages. J. Virol. Methods 2000, 86, 13–20. [Google Scholar] [CrossRef]
- Singh, I.; McConnell, I.; Blacklaws, B. Immune response to individual Maedi-Visna Virus gag antigens. J. Virol. 2006, 80, 912–919. [Google Scholar] [CrossRef]
- Fluri, A.; Nenci, C.; Zahno, M.-L.; Vogt, H.; Charan, S.; Busato, A.; Pancino, G.; Peterhans, E.; Obexer-Ruff, G.; Bertoni, G. The MHC-haplotype influences primary, but not memory, immune responses to an immunodominant peptide containing T- and B-cell epitopes of the caprine arthritis encephalitis virus Gag protein. Vaccine 2006, 24, 597–606. [Google Scholar] [CrossRef]
- Clements, J.E.; Zink, M.C. Molecular biology and pathogenesis of animal lentivirus infections. Clin. Microbiol. Rev. 1996, 9, 100–117. [Google Scholar]
- Staskus, K.A.; Couch, L.; Bitterman, P.; Retzel, E.F.; Zupancic, M.; List, J.; Haase, A.T. In situ amplification of visna virus DNA in tissue sections reveals a reservoir of latently infected cells. Microb. Pathog. 1991, 11, 67–76. [Google Scholar]
- Reina, R.; Glaria, I.; Benavides, J.; de Andrés, X.; Crespo, H.; Solano, C.; Pérez, V.; Luján, L.; Pérez, M.M.; Pérez de la Lastra, J.M.; et al. Association of CD80 and CD86 expression levels with disease status of Visna/Maedi virus infected sheep. Viral Immunol. 2007, 20, 609–622. [Google Scholar] [CrossRef]
- Gudnadóttir, M.; Demosthenous, A.; Hadjisavvas, T. Vaccination delays Maedi-Visna lentivirus infection in a naturally-infected sheep flock. BMC Vet. Res. 2013, 9, e16. [Google Scholar] [CrossRef]
- Benavides, J.; García-Pariente, C.; Ferreras, M.C.; Fuertes, M.; García-Marín, J.F.; Pérez, V. Diagnosis of clinical cases of the nervous form of Maedi-Visna in 4- and 6-month-old lambs. Vet. J. 2007, 174, 255–258. [Google Scholar]
- Brodie, S.J.; Pearson, L.D.; Zink, M.C.; Bickle, H.M.; Anderson, B.C.; Marcom, K.A.; Demartinit, J.C.; DeMartini, J.C. Ovine lentivirus expression and disease. Virus replication, but not entry, is restricted to macrophages of specific tissues. Am. J. Pathol. 1995, 146, 250–263. [Google Scholar]
- Agnarsdóttir, G.; Thorsteinsdóttir, H.; Oskarsson, T.; Matthíasdóttir, S.; Haflidadóttir, B.S.; Andrésson, O.S.; Andrésdóttir, V. The long terminal repeat is a determinant of cell tropism of Maedi-Visna Virus. J. Gen. Virol. 2000, 81, 1901–1905. [Google Scholar]
- Hötzel, I.; Cheevers, W.P. A maedi-visna virus strain K1514 receptor gene is located in sheep chromosome 3p and the syntenic region of human chromosome 2. J. Gen. Virol. 2002, 83, 1759–1764. [Google Scholar]
- Hötzel, I.; Cheevers, W.P. Host range of small-ruminant lentivirus cytopathic variants determined with a selectable caprine arthritis-encephalitis virus pseudotype system. J. Virol. 2001, 75, 7384–7391. [Google Scholar] [CrossRef]
- Crespo, H.; Jauregui, P.; Glaria, I.; José, I.S.; Polledo, L.; García-Marín, J.F.; Luján, L.; de Andrés, D.; Amorena, B.; Reina, R. Mannose receptor may be involved in small ruminant lentivirus pathogenesis. Vet. Res. 2012, 43, e43. [Google Scholar] [CrossRef] [Green Version]
- Amills, M.; Ramiya, V.; Norimine, J.; Lewin, H.A. The major histocompatibility complex of ruminants. Rev. Sci. Tech. 1998, 17, 108–120. [Google Scholar]
- Dukkipati, V.S.R.; Blair, H.T.; Garrick, D.J.; Murray, A. “Ovar-Mhc”—Ovine major histocompatibility complex: Structure and gene polymorphisms. Genet. Mol. Res. 2006, 5, 581–608. [Google Scholar]
- Sayers, G.; Sweeney, T. Gastrointestinal nematode infection in sheep—A review of the alternatives to anthelmintics in parasite control. Anim. Health Res. Rev. 2005, 6, 159–172. [Google Scholar] [CrossRef]
- Lee, W.C.; Bird, P.; McConnell, I.; Watt, N.J.; Blacklaws, B. The phenotype and phagocytic activity of macrophages during Maedi-Visna Virus infection. Vet. Immunol. Immunopathol. 1996, 51, 113–126. [Google Scholar] [CrossRef]
- Bergsteinsdóttir, K.; Arnadóttir, S.; Torsteinsdóttir, S.; Agnarsdóttir, G.; Andrésdóttir, V.; Péttursson, G.; Georgsson, G. Constitutive and visna virus induced expression of class I and II major histocompatibility complex antigens in the central nervous system of sheep and their role in the pathogenesis of visna lesions. Neuropathol. Appl. Neurobiol. 1998, 24, 224–232. [Google Scholar] [CrossRef]
- Harkiss, G.D.; Watt, N.J.; King, T.J.; Williams, J.; Hopkins, J. Retroviral arthritis: Phenotypic analysis of cells in the synovial fluid of sheep with inflammatory synovitis associated with visna virus infection. Clin. Immunol. Immunopathol. 1991, 60, 106–117. [Google Scholar] [CrossRef]
- Anderson, A.A.; Harkiss, G.D.; Watt, N.J. Quantitative analysis of immunohistological changes in the synovial membrane of sheep infected with Maedi-Visna Virus. Clin. Immunol. Immunopathol. 1994, 72, 21–29. [Google Scholar] [CrossRef]
- Larruskain, A.; Minguijón, E.; García-etxebarria, K.; Moreno, B.; Arostegui, I.; Juste, R.A; Jugo, B.M. MHC class II DRB1 gene polymorphism in the pathogenesis of Maedi-Visna and pulmonary adenocarcinoma viral diseases in sheep. Immunogenetics 2010, 62, 75–83. [Google Scholar] [CrossRef]
- Larruskain, A.; Minguijón, E.; Arostegui, I.; Moreno, B.; Juste, R.A.; Jugo, B.M. Microsatellites in immune-relevant regions and their associations with Maedi-Visna and ovine pulmonary adenocarcinoma viral diseases. Vet. Immunol. Immunopathol. 2012, 145, 438–446. [Google Scholar] [CrossRef]
- Ruff, G.; Regli, J.G.; Lazary, S. Occurrence of caprine leucocyte class I and II antigens in Saanen goats affected by caprine arthritis (CAE). Eur. J. Immunogenet. 1993, 20, 285–288. [Google Scholar] [CrossRef]
- Batalia, M.A.; Collins, E.J.; Carolina, N. Peptide binding by class I and class II MHC molecules. Biopolymers 1997, 43, 281–302. [Google Scholar] [CrossRef]
- Konnai, S.; Takeshima, S.; Tajima, S.; Yin, S.A.; Okada, K.; Onuma, M.; Aida, Y. The influence of ovine MHC class II DRB1 alleles on immune response in bovine leukemia virus infection. Microbiol. Immunol. 2003, 47, 223–232. [Google Scholar]
- De Andrés, D.; Klein, D.; Watt, N.J.; Berriatua, E.; Torsteinsdottir, S.; Blacklaws, B.A.; Harkiss, G.D. Diagnostic tests for small ruminant lentiviruses. Vet. Microbiol. 2005, 107, 49–62. [Google Scholar] [CrossRef]
- Singh, D.K.; Chebloune, Y.; Mselli-Lakhal, L.; Karr, B.M.; Narayan, O. Ovine lentivirus-infected macrophages mediate productive infection in cell types that are not susceptible to infection with cell-free virus. J. Gen. Virol. 1999, 80, 1437–1444. [Google Scholar]
- Torsteinsdóttir, S.; Georgsson, G.; Gísladóttir, E.; Rafnar, B.; Pálsson, P.A.; Pétursson, G. Pathogenesis of central nervous system lesions in visna: Cell-mediated immunity and lymphocyte subsets in blood, brain and cerebrospinal fluid. J. Neuroimmunol. 1992, 41, 149–158. [Google Scholar] [CrossRef]
- Perry, L.L.; Wilkerson, M.J.; Hullinger, G.A.; Cheevers, W.P. Depressed CD4T lymphocyte proliferative response and enhanced antibody response to viral antigen in chronic lentivirus-induced arthritis. J. Infect. Dis. 1995, 171, 328–334. [Google Scholar] [CrossRef]
- Trujillo, J.D.; Hotzel, K.J.; Snekvik, K.R.; Cheevers, W.P. Antibody response to the surface envelope of caprine arthritis-encephalitis lentivirus: Disease status is predicted by SU antibody isotype. Virology 2004, 325, 129–136. [Google Scholar] [CrossRef]
- Bird, P.; Reyburn, H.T.; Blacklaws, B.A.; Allen, D.; Nettleton, P.; Yirrell, D.L.; Watt, N.; Sargan, D.; McConnell, I. The restricted IgG1 antibody response to maedi visna virus is seen following infection but not following immunization with recombinant gag protein. Clin. Exp. Immunol. 1995, 102, 274–280. [Google Scholar]
- Watt, N.J.; MacIntyre, N.; Collie, D.; Sargan, D.; McConnell, I. Phenotypic analysis of lymphocyte populations in the lungs and regional lymphoid tissue of sheep naturally infected with Maedi-Visna Virus. Clin. Exp. Immunol. 1992, 90, 204–208. [Google Scholar]
- Luján, L.; Begara, I.; Collie, D.D.; Watt, N.J. CD8+ lymphocytes in bronchoalveolar lavage and blood: In vivo indicators of lung pathology caused by Maedi-Visna Virus. Vet. Immunol. Immunopathol. 1995, 49, 89–100. [Google Scholar] [CrossRef]
- Jolly, P.E.; Gangopadhyay, A.; Chen, S.; Reddy, P.G.; Weiss, H.L. Changes in the leukocyte phenotype profile of goats infected with the caprine arthritis encephalitis virus. Vet. Immunol. Immunopathol. 1997, 56, 97–106. [Google Scholar] [CrossRef]
- Blacklaws, B.A; Bird, P.; Allen, D.; McConnell, I. Circulating cytotoxic T lymphocyte precursors in Maedi-Visna Virus-infected sheep. J. Gen. Virol. 1994, 75, 1589–1596. [Google Scholar] [CrossRef]
- Lee, W.C.; McConnell, I.; Blacklaws, B.A. Cytotoxic activity against Maedi-Visna Virus-infected macrophages. J. Virol. 1994, 68, 8331–8338. [Google Scholar]
- Estes, D.M.; Brown, W.C. Type 1 and type 2 responses in regulation of Ig isotype expression in cattle. Vet. Immunol. Immunopathol. 2002, 90, 1–10. [Google Scholar] [CrossRef]
- Snekvik, K.R.; Beyer, J.C.; Bertoni, G.; von Beust, B.R.; Baszler, T.V; Palmer, G.H.; McElwain, T.F.; Cheevers, W.P. Characterization of caprine interleukin-4. Vet. Immunol. Immunopathol. 2001, 78, 219–229. [Google Scholar] [CrossRef]
- Lechner, F.; Vogt, H.R.; Seow, H.F.; Bertoni, G.; Cheevers, W.P.; von Bodungen, U.; Zurbriggen, A; Peterhans, E. Expression of cytokine mRNA in lentivirus-induced arthritis. Am. J. Pathol. 1997, 151, 1053–1065. [Google Scholar]
- Woodall, C.J.; Maclaren, L.J.; Watt, N.J. Differential levels of mRNAs for cytokines, the interleukin-2 receptor and Class II DR/DQ genes in ovine interstitial pneumonia induced by Maedi visna virus infection. Vet. Pathol. 1997, 34, 204–211. [Google Scholar] [CrossRef]
- Legastelois, I.; Cottin, V.; Mornex, J.F.; Cordier, G. Alveolar macrophages from sheep naturally infected by Visna-Maedi virus contribute to IL-8 production in the lung. Vet. Immunol. Immunopathol. 1997, 59, 131–139. [Google Scholar] [CrossRef]
- Zhang, Z.; Harkiss, G.D.; Hopkins, J. Granulocyte macrophage colony stimulating factor is elevated in alveolar macrophages from sheep naturally infected with Maedi-Visna Virus and stimulates Maedi-Visna Virus replication in macrophages in vitro. Clin. Exp. Immunol. 2002, 129, 240–246. [Google Scholar] [CrossRef]
- Legastelois, I.; Cordier, O.; Cozon, G.; Cador, J. Visna-Maedi virus-induced expression of interleukin-8 gene in sheep alveolar cells following experimental in vitro and in vivo infection. Res. Virol. 1996, 147, 191–197. [Google Scholar] [CrossRef]
- Ellis, J.A.; DeMartini, J.C. Ovine interleukin-2: Partial purification and assay in normal sheep and sheep with ovine progressive pneumonia. Vet. Immunol. Immunopathol. 1985, 8, 15–25. [Google Scholar] [CrossRef]
- Legastelois, I.; Levrey, H.; Greenland, T.; Mornex, J.F.; Cordier, G. Visna-Maedi virus induces interleukin-8 in sheep alveolar macrophages through a tyrosine-kinase signaling pathway. Am. J. Respir. Cell Mol. Biol. 1998, 18, 532–537. [Google Scholar] [CrossRef]
- Larruskain, A.; Bernales, I.; Luján, L.; de Andrés, D.; Amorena, B.; Jugo, B.M. Expression analysis of 13 ovine immune response candidate genes in Visna/Maedi disease progression. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 405–413. [Google Scholar] [CrossRef]
- White, S.N.; Mousel, M.R.; Reynolds, J.O.; Lewis, G.S.; Herrmann-Hoesing, L.M. Common promoter deletion is associated with 3.9-fold differential transcription of ovine CCR5 and reduced proviral level of ovine progressive pneumonia virus. Anim. Genet. 2009, 40, 583–589. [Google Scholar] [CrossRef]
- Shirakawa, I.; Deichmann, K.A.; Izuhara, I.; Mao, I.; Adra, C.N.; Hopkin, J.M. Atopy and asthma: Genetic variants of IL-4 and IL-13 signalling. Immunol. Today 2000, 21, 60–64. [Google Scholar] [CrossRef]
- Locati, M.; Otero, K.; Schioppa, T.; Signorelli, P.; Perrier, P.; Baviera, S.; Sozzani, S.; Mantovani, A. The chemokine system: Tuning and shaping by regulation of receptor expression and coupling in polarized responses. Allergy 2002, 57, 972–982. [Google Scholar] [CrossRef]
- Medzhitov, R. Recognition of microorganisms and activation of the immune response. Nature 2007, 449, 819–826. [Google Scholar] [CrossRef]
- Werling, D.; Jann, O.; Offord, V.; Glass, E.; Coffey, T. Variation matters: TLR structure and species-specific pathogen recognition. Trends mmunol. 2009, 30, 124–130. [Google Scholar] [CrossRef]
- Takeda, K.; Kaisho, T.; Akira, S. Toll-like receptors. Annu. Rev. Immunol. 2003, 21, 335–376. [Google Scholar] [CrossRef]
- Mikula, I.; Bhide, M.; Pastorekova, S. Characterization of ovine TLR7 and TLR8 protein coding regions, detection of mutations and Maedi-Visna Virus infection. Vet. Immunol. Immunopathol. 2010, 138, 51–59. [Google Scholar] [CrossRef]
- Bell, J.K.; Mullen, G.E.; Leifer, C.A.; Mazzoni, A.; Davies, D.R.; Segal, D.M. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003, 24, 528–533. [Google Scholar] [CrossRef]
- Zheng, Y.H.; Peterlin, B.B. Intracellular immunity to HIV-1: Newly defined retroviral battles inside infected cells. Retrovirology 2005, 2, e25. [Google Scholar] [CrossRef] [Green Version]
- Huthoff, H.; Towers, G.J. Restriction of retroviral replication by APOBEC3G/F and TRIM5alpha. Trends Microbiol. 2008, 16, 612–619. [Google Scholar] [CrossRef]
- Jáuregui, P.; Crespo, H.; Glaria, I.; Luján, L.; Contreras, A.; Rosati, S.; de Andrés, D.; Amorena, B.; Towers, G.J.; Reina, R. Ovine TRIM5αcan restrict Visna/Maedi virus. J. Virol. 2012, 86, 9504–9509. [Google Scholar] [CrossRef]
- Van Manen, D.; Rits, M.A.; Beugeling, C.; van Dort, K.; Schuitemaker, H.; Kootstra, N.A. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog. 2008, 4, e18. [Google Scholar] [CrossRef]
- LaRue, R.S.; Jónsson, S.R.; Silverstein, K.A.T.; Lajoie, M.; Bertrand, D.; El-Mabrouk, N.; Hötzel, I.; Andrésdóttir, V.; Smith, T.P.L.; Harris, R.S. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 2008, 9, 10–1186. [Google Scholar]
- LaRue, R.S.; Lengyel, J.; Jonsson, S.R.; Andresdottir, V.; Harris, R.S. Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J. Virol. 2010, 84, 8193–8201. [Google Scholar] [CrossRef]
- Jónsson, S.R.; Haché, G.; Stenglein, M.D.; Fahrenkrug, S.C.; Andrésdóttir, V.; Harris, R.S. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 2006, 34, 5683–5694. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Larruskain, A.; Jugo, B.M. Retroviral Infections in Sheep and Goats: Small Ruminant Lentiviruses and Host Interaction. Viruses 2013, 5, 2043-2061. https://doi.org/10.3390/v5082043
Larruskain A, Jugo BM. Retroviral Infections in Sheep and Goats: Small Ruminant Lentiviruses and Host Interaction. Viruses. 2013; 5(8):2043-2061. https://doi.org/10.3390/v5082043
Chicago/Turabian StyleLarruskain, Amaia, and Begoña M. Jugo. 2013. "Retroviral Infections in Sheep and Goats: Small Ruminant Lentiviruses and Host Interaction" Viruses 5, no. 8: 2043-2061. https://doi.org/10.3390/v5082043




