From Crescent to Mature Virion: Vaccinia Virus Assembly and Maturation
Abstract
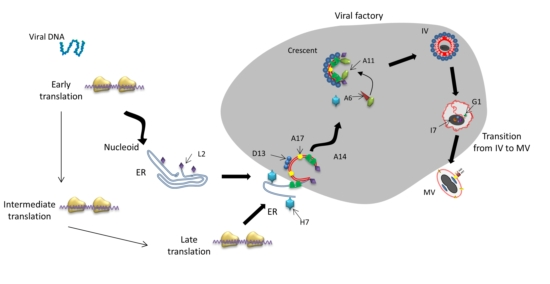
:1. Introduction
2. Entry of Vaccinia into Host Cells
3. Viral Factory Formation and Function
4. Crescent Formation
4.1. Essential Structural Proteins for Crescent Formation: D13, A14 and A17
4.1.1. The Scaffold of Crescent Membranes—D13
4.1.2. Key Proteins Anchored in the Virion Membrane—A14 and A17
4.2. Regulatory Proteins that are Essential for Viral Membrane Assembly
4.2.1. Early Transcribed Membrane Regulatory Protein—L2
4.2.2. A Small, L2-Interacting Protein—A30.5
4.2.3. A11, a Viral Factory-Located, and Membrane-Associated Protein
4.2.4. Trafficking Protein A6
4.2.5. H7, a Cytoplasmic Located Protein
4.2.6. Major Protein Kinase in the Virion Core—F10

5. Immature Virion Formation
5.1. Proteins Involved in Viroplasm-Membrane Association
5.1.1. The Seven Protein Complex
5.1.2. E6 Is Also Involved in Membrane-Viroplasm Association
5.2. How the Genome Becomes Incorporated into the Virion Remains Unclear
6. Transition of Immature Virus Particles to Mature Virions
6.1. Removal of the D13 Scaffold is Necessary but Not Sufficient for the Transition of IVs to MVs
6.2. Membrane Restructuring to Form MVs Remains Controversial
6.3. Transcriptional Activity May Be Linked to a Stable Core Structure
6.4. Morphogenic Proteolysis Generates an Active Core
6.5. Cellular Host Proteins Can Have Roles in Morphogenesis
6.6. The VACV Redox System is Important for Functional Membrane Proteins Displayed on the MV Surface
6.7. Lateral Bodies Package Viral Proteins in Preparation for Delivery to the Cytosol of Host Cells

7. A Subset of MVs Go on to Form Enveloped Virions
8. Conclusions and Open Questions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jacobs, B.L.; Langland, J.O.; Kibler, K.V.; Denzler, K.L.; White, S.D.; Holechek, S.A.; Wong, S.; Huynh, T.; Baskin, C.R. Vaccinia virus vaccines: Past, present and future. Antivir. Res. 2009, 84, 1–13. [Google Scholar]
- Walsh, S.R.; Dolin, R. Vaccinia viruses: Vaccines against smallpox and vectors against infectious diseases and tumors. Expert Rev. Vaccines 2011, 10, 1221–1240. [Google Scholar]
- Johnson, G.P.; Goebel, S.J.; Paoletti, E. An update on the vaccinia virus genome. Virology 1993, 196, 381–401. [Google Scholar]
- Goebel, S.J.; Johnson, G.P.; Perkus, M.E.; Davis, S.W.; Winslow, J.P.; Paoletti, E. The complete DNA sequence of vaccinia virus. Virology 1990, 179, 247–266, 517–563. [Google Scholar]
- Condit, R.C.; Moussatche, N.; Traktman, P. In a nutshell: Structure and assembly of the vaccinia virion. Adv. Virus Res. 2006, 66, 31–124. [Google Scholar]
- Roberts, K.L.; Smith, G.L. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 2008, 16, 472–479. [Google Scholar]
- Prichard, M.N.; Kern, E.R. Orthopoxvirus targets for the development of new antiviral agents. Antivir. Res. 2012, 94, 111–125. [Google Scholar]
- Townsley, A.C.; Weisberg, A.S.; Wagenaar, T.R.; Moss, B. Vaccinia virus entry into cells via a low-pH-dependent endosomal pathway. J. Virol. 2006, 80, 8899–8908. [Google Scholar]
- Townsley, A.C.; Moss, B. Two distinct low-pH steps promote entry of vaccinia virus. J. Virol. 2007, 81, 8613–8620. [Google Scholar]
- Schmidt, F.I.; Bleck, C.K.; Mercer, J. Poxvirus host cell entry. Curr. Opin. Virol. 2012, 2, 20–27. [Google Scholar]
- Hsiao, J.C.; Chung, C.S.; Chang, W. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: Identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J. Virol. 1998, 72, 8374–8379. [Google Scholar]
- Chung, C.S.; Hsiao, J.C.; Chang, Y.S.; Chang, W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 1998, 72, 1577–1585. [Google Scholar]
- Hsiao, J.C.; Chung, C.S.; Chang, W. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 1999, 73, 8750–8761. [Google Scholar]
- Lin, C.L.; Chung, C.S.; Heine, H.G.; Chang, W. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 2000, 74, 3353–3365. [Google Scholar]
- Vazquez, M.I.; Esteban, M. Identification of functional domains in the 14-kilodalton envelope protein (A27L) of vaccinia virus. J. Virol. 1999, 73, 9098–9109. [Google Scholar]
- Satheshkumar, P.S.; Chavre, J.; Moss, B. Role of the vaccinia virus O3 protein in cell entry can be fulfilled by its Sequence flexible transmembrane domain. Virology 2013, 444, 148–157. [Google Scholar]
- Satheshkumar, P.S.; Moss, B. Sequence-divergent chordopoxvirus homologs of the o3 protein maintain functional interactions with components of the vaccinia virus entry-fusion complex. J. Virol. 2012, 86, 1696–1705. [Google Scholar]
- Senkevich, T.G.; Ward, B.M.; Moss, B. Vaccinia virus entry into cells is dependent on a virion surface protein encoded by the A28L gene. J. Virol. 2004, 78, 2357–2366. [Google Scholar]
- Townsley, A.C.; Senkevich, T.G.; Moss, B. Vaccinia virus A21 virion membrane protein is required for cell entry and fusion. J. Virol. 2005, 79, 9458–9469. [Google Scholar]
- Townsley, A.C.; Senkevich, T.G.; Moss, B. The product of the vaccinia virus L5R gene is a fourth membrane protein encoded by all poxviruses that is required for cell entry and cell-cell fusion. J. Virol. 2005, 79, 10988–10998. [Google Scholar]
- Ojeda, S.; Senkevich, T.G.; Moss, B. Entry of vaccinia virus and cell-cell fusion require a highly conserved cysteine-rich membrane protein encoded by the A16L gene. J. Virol. 2006, 80, 51–61. [Google Scholar]
- Turner, P.C.; Dilling, B.P.; Prins, C.; Cresawn, S.G.; Moyer, R.W.; Condit, R.C. Vaccinia virus temperature-sensitive mutants in the A28 gene produce non-infectious virions that bind to cells but are defective in entry. Virology 2007, 366, 62–72. [Google Scholar]
- Izmailyan, R.A.; Huang, C.Y.; Mohammad, S.; Isaacs, S.N.; Chang, W. The envelope G3L protein is essential for entry of vaccinia virus into host cells. J. Virol. 2006, 80, 8402–8410. [Google Scholar]
- Nelson, G.E.; Wagenaar, T.R.; Moss, B. A conserved sequence within the H2 subunit of the vaccinia virus entry/fusion complex is important for interaction with the A28 subunit and infectivity. J. Virol. 2008, 82, 6244–6250. [Google Scholar]
- Senkevich, T.G.; Moss, B. Vaccinia virus H2 protein is an essential component of a complex involved in virus entry and cell-cell fusion. J. Virol. 2005, 79, 4744–4754. [Google Scholar]
- Ojeda, S.; Domi, A.; Moss, B. Vaccinia virus G9 protein is an essential component of the poxvirus entry-fusion complex. J. Virol. 2006, 80, 9822–9830. [Google Scholar]
- Chang, S.J.; Shih, A.C.; Tang, Y.L.; Chang, W. Vaccinia mature virus fusion regulator A26 protein binds to A16 and G9 proteins of the viral entry fusion complex and dissociates from mature virions at low pH. J. Virol. 2012, 86, 3809–3818. [Google Scholar]
- Brown, E.; Senkevich, T.G.; Moss, B. Vaccinia virus F9 virion membrane protein is required for entry but not virus assembly, in contrast to the related L1 protein. J. Virol. 2006, 80, 9455–9464. [Google Scholar]
- Bisht, H.; Weisberg, A.S.; Moss, B. Vaccinia virus l1 protein is required for cell entry and membrane fusion. J. Virol. 2008, 82, 8687–8694. [Google Scholar]
- Moss, B. Poxvirus cell entry: How many proteins does it take? Viruses 2012, 4, 688–707. [Google Scholar]
- Tolonen, N.; Doglio, L.; Schleich, S.; Krijnse Locker, J. Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Mol. Biol. Cell 2001, 12, 2031–2046. [Google Scholar]
- Cepeda, V.; Esteban, M. Novel insights on the progression of intermediate viral forms in the morphogenesis of vaccinia virus. Virus Res. 2014, 183, 23–29. [Google Scholar]
- Schramm, B.; Locker, J.K. Cytoplasmic organization of POXvirus DNA replication. Traffic 2005, 6, 839–846. [Google Scholar]
- De Castro, I.F.; Volonte, L.; Risco, C. Virus factories: Biogenesis and structural design. Cell. Microbiol. 2013, 15, 24–34. [Google Scholar]
- Katsafanas, G.C.; Moss, B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2007, 2, 221–228. [Google Scholar]
- Chlanda, P.; Carbajal, M.A.; Cyrklaff, M.; Griffiths, G.; Krijnse-Locker, J. Membrane rupture generates single open membrane sheets during vaccinia virus assembly. Cell Host Microbe 2009, 6, 81–90. [Google Scholar]
- Meng, X.; Wu, X.; Yan, B.; Deng, J.; Xiang, Y. Analysis of the role of vaccinia virus H7 in virion membrane biogenesis with an H7-deletion mutant. J. Virol. 2013, 87, 8247–8253. [Google Scholar]
- Dales, S.; Mosbach, E.H. Vaccinia as a model for membrane biogenesis. Virology 1968, 35, 564–583. [Google Scholar]
- Risco, C.; Rodriguez, J.R.; Lopez-Iglesias, C.; Carrascosa, J.L.; Esteban, M.; Rodriguez, D. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 2002, 76, 1839–1855. [Google Scholar]
- Hollinshead, M.; Vanderplasschen, A.; Smith, G.L.; Vaux, D.J. Vaccinia virus intracellular mature virions contain only one lipid membrane. J. Virol. 1999, 73, 1503–1517. [Google Scholar]
- Szajner, P.; Weisberg, A.S.; Lebowitz, J.; Heuser, J.; Moss, B. External scaffold of spherical immature poxvirus particles is made of protein trimers, forming a honeycomb lattice. J. Cell Biol. 2005, 170, 971–981. [Google Scholar]
- Sodeik, B.; Krijnse-Locker, J. Assembly of vaccinia virus revisited: De novo membrane synthesis or acquisition from the host? Trends Microbiol. 2002, 10, 15–24. [Google Scholar]
- Husain, M.; Weisberg, A.S.; Moss, B. Existence of an operative pathway from the endoplasmic reticulum to the immature poxvirus membrane. Proc. Natl. Acad. Sci. USA 2006, 103, 19506–19511. [Google Scholar]
- Maruri-Avidal, L.; Weisberg, A.S.; Moss, B. Vaccinia virus L2 protein associates with the endoplasmic reticulum near the growing edge of crescent precursors of immature virions and stabilizes a subset of viral membrane proteins. J. Virol. 2011, 85, 12431–12441. [Google Scholar]
- Maruri-Avidal, L.; Weisberg, A.S.; Bisht, H.; Moss, B. Analysis of viral membranes formed in cells infected by a vaccinia virus L2-deletion mutant suggests their origin from the endoplasmic reticulum. J. Virol. 2013, 87, 1861–1871. [Google Scholar]
- Maruri-Avidal, L.; Weisberg, A.S.; Moss, B. Direct formation of vaccinia virus membranes from the endoplasmic reticulum in the absence of the newly characterized L2-interacting protein A30.5. J. Virol. 2013, 87, 12313–12326. [Google Scholar]
- Suarez, C.; Welsch, S.; Chlanda, P.; Hagen, W.; Hoppe, S.; Kolovou, A.; Pagnier, I.; Raoult, D.; Krijnse Locker, J. Open membranes are the precursors for assembly of large DNA viruses. Cell. Microbiol. 2013, 15, 1883–1895. [Google Scholar]
- Heuser, J. Deep-etch EM reveals that the early poxvirus envelope is a single membrane bilayer stabilized by a geodetic “honeycomb” surface coat. J. Cell Biol. 2005, 169, 269–283. [Google Scholar]
- Bahar, M.W.; Graham, S.C.; Stuart, D.I.; Grimes, J.M. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure 2011, 19, 1011–1020. [Google Scholar]
- Zhang, Y.; Moss, B. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology 1992, 187, 643–653. [Google Scholar]
- Sodeik, B.; Griffiths, G.; Ericsson, M.; Moss, B.; Doms, R.W. Assembly of vaccinia virus: Effects of rifampin on the intracellular distribution of viral protein p65. J. Virol. 1994, 68, 1103–1114. [Google Scholar]
- Mohandas, A.R.; Dales, S. Involvement of spicules in the formation of vaccinia virus envelopes elucidated by a conditional lethal mutant. Virology 1995, 214, 494–502. [Google Scholar]
- Charity, J.C.; Katz, E.; Moss, B. Amino acid substitutions at multiple sites within the vaccinia virus D13 scaffold protein confer resistance to rifampicin. Virology 2007, 359, 227–232. [Google Scholar]
- Baldick, C.J., Jr.; Moss, B. Resistance of vaccinia virus to rifampicin conferred by a single nucleotide substitution near the predicted NH2 terminus of a gene encoding an Mr 62,000 polypeptide. Virology 1987, 156, 138–145. [Google Scholar]
- Hyun, J.K.; Accurso, C.; Hijnen, M.; Schult, P.; Pettikiriarachchi, A.; Mitra, A.K.; Coulibaly, F. Membrane remodeling by the double-barrel scaffolding protein of poxvirus. PLoS Pathog. 2011, 7, e1002239. [Google Scholar]
- Bisht, H.; Weisberg, A.S.; Szajner, P.; Moss, B. Assembly and disassembly of the capsid-like external scaffold of immature virions during vaccinia virus morphogenesis. J. Virol. 2009, 83, 9140–9150. [Google Scholar]
- Betakova, T.; Wolffe, E.J.; Moss, B. Membrane topology of the vaccinia virus A17L envelope protein. Virology 1999, 261, 347–356. [Google Scholar]
- Krijnse-Locker, J.; Schleich, S.; Rodriguez, D.; Goud, B.; Snijder, E.J.; Griffiths, G. The role of a 21-kDa viral membrane protein in the assembly of vaccinia virus from the intermediate compartment. J. Biol. Chem. 1996, 271, 14950–14958. [Google Scholar]
- Unger, B.; Mercer, J.; Boyle, K.A.; Traktman, P. Biogenesis of the vaccinia virus membrane: Genetic and ultrastructural analysis of the contributions of the A14 and A17 proteins. J. Virol. 2013, 87, 1083–1097. [Google Scholar]
- Mercer, J.; Traktman, P. Investigation of structural and functional motifs within the vaccinia virus A14 phosphoprotein, an essential component of the virion membrane. J. Virol. 2003, 77, 8857–8871. [Google Scholar]
- Traktman, P.; Liu, K.; DeMasi, J.; Rollins, R.; Jesty, S.; Unger, B. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: Construction and characterization of a tetracycline-inducible recombinant. J. Virol. 2000, 74, 3682–3695. [Google Scholar]
- Wallengren, K.; Risco, C.; Krijnse-Locker, J.; Esteban, M.; Rodriguez, D. The A17L gene product of vaccinia virus is exposed on the surface of IMV. Virology 2001, 290, 143–152. [Google Scholar]
- Betakova, T.; Wolffe, E.J.; Moss, B. Regulation of vaccinia virus morphogenesis: Phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L kinase. J. Virol. 1999, 73, 3534–3543. [Google Scholar]
- Derrien, M.; Punjabi, A.; Khanna, M.; Grubisha, O.; Traktman, P. Tyrosine phosphorylation of A17 during vaccinia virus infection: Involvement of the H1 phosphatase and the F10 kinase. J. Virol. 1999, 73, 7287–7296. [Google Scholar]
- Rodriguez, J.R.; Risco, C.; Carrascosa, J.L.; Esteban, M.; Rodriguez, D. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J. Virol. 1998, 72, 1287–1296. [Google Scholar]
- Rodriguez, D.; Esteban, M.; Rodriguez, J.R. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J. Virol. 1995, 69, 4640–4648. [Google Scholar]
- Rodriguez, D.; Risco, C.; Rodriguez, J.R.; Carrascosa, J.L.; Esteban, M. Inducible expression of the vaccinia virus A17L gene provides a synchronized system to monitor sorting of viral proteins during morphogenesis. J. Virol. 1996, 70, 7641–7653. [Google Scholar]
- Wolffe, E.J.; Moore, D.M.; Peters, P.J.; Moss, B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J. Virol. 1996, 70, 2797–2808. [Google Scholar]
- Chlanda, P.; Carbajal, M.A.; Kolovou, A.; Hamasaki, M.; Cyrklaff, M.; Griffiths, G.; Krijnse-Locker, J. Vaccinia virus lacking A17 induces complex membrane structures composed of open membrane sheets. Arch. Virol. 2011, 156, 1647–1653. [Google Scholar]
- Maruri-Avidal, L.; Domi, A.; Weisberg, A.S.; Moss, B. Participation of vaccinia virus l2 protein in the formation of crescent membranes and immature virions. J. Virol. 2011, 85, 2504–2511. [Google Scholar]
- Resch, W.; Weisberg, A.S.; Moss, B. Vaccinia virus nonstructural protein encoded by the A11R gene is required for formation of the virion membrane. J. Virol. 2005, 79, 6598–6609. [Google Scholar]
- Maruri-Avidal, L.; Weisberg, A.S.; Moss, B. Association of the vaccinia virus A11 protein with the endoplasmic reticulum and crescent precursors of immature virions. J. Virol. 2013, 87, 10195–10206. [Google Scholar]
- Wu, X.; Meng, X.; Yan, B.; Rose, L.; Deng, J.; Xiang, Y. Vaccinia virus virion membrane biogenesis protein A11 associates with viral membranes in a manner that requires the expression of another membrane biogenesis protein, A6. J. Virol. 2012, 86, 11276–11286. [Google Scholar]
- Meng, X.; Embry, A.; Sochia, D.; Xiang, Y. Vaccinia virus A6L encodes a virion core protein required for formation of mature virion. J. Virol. 2007, 81, 1433–1443. [Google Scholar]
- Meng, X.; Embry, A.; Rose, L.; Yan, B.; Xu, C.; Xiang, Y. Vaccinia virus A6 is essential for virion membrane biogenesis and localization of virion membrane proteins to sites of virion assembly. J. Virol. 2012, 86, 5603–5613. [Google Scholar]
- Satheshkumar, P.S.; Weisberg, A.; Moss, B. Vaccinia virus H7 protein contributes to the formation of crescent membrane precursors of immature virions. J. Virol. 2009, 83, 8439–8450. [Google Scholar]
- Traktman, P.; Caligiuri, A.; Jesty, S.A.; Liu, K.; Sankar, U. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of virion morphogenesis. J. Virol. 1995, 69, 6581–6587. [Google Scholar]
- Wang, S.; Shuman, S. Vaccinia virus morphogenesis is blocked by temperature-sensitive mutations in the F10 gene, which encodes protein kinase 2. J. Virol. 1995, 69, 6376–6388. [Google Scholar]
- Lin, S.; Broyles, S.S. Vaccinia protein kinase 2: A second essential serine/threonine protein kinase encoded by vaccinia virus. Proc. Natl. Acad. Sci. USA 1994, 91, 7653–7657. [Google Scholar]
- Punjabi, A.; Traktman, P. Cell biological and functional characterization of the vaccinia virus F10 kinase: Implications for the mechanism of virion morphogenesis. J. Virol. 2005, 79, 2171–2190. [Google Scholar]
- Szajner, P.; Weisberg, A.S.; Moss, B. Evidence for an essential catalytic role of the F10 protein kinase in vaccinia virus morphogenesis. J. Virol. 2004, 78, 257–265. [Google Scholar]
- Morgan, C. The insertion of DNA into vaccinia virus. Science 1976, 193, 591–592. [Google Scholar]
- Szajner, P.; Weisberg, A.S.; Wolffe, E.J.; Moss, B. Vaccinia virus A30L protein is required for association of viral membranes with dense viroplasm to form immature virions. J. Virol. 2001, 75, 5752–5761. [Google Scholar]
- Szajner, P.; Jaffe, H.; Weisberg, A.S.; Moss, B. Vaccinia virus G7L protein Interacts with the A30L protein and is required for association of viral membranes with dense viroplasm to form immature virions. J. Virol. 2003, 77, 3418–3429. [Google Scholar]
- Szajner, P.; Jaffe, H.; Weisberg, A.S.; Moss, B. A complex of seven vaccinia virus proteins conserved in all chordopoxviruses is required for the association of membranes and viroplasm to form immature virions. Virology 2004, 330, 447–459. [Google Scholar]
- Mercer, J.; Traktman, P. Genetic and cell biological characterization of the vaccinia virus A30 and G7 phosphoproteins. J. Virol. 2005, 79, 7146–7161. [Google Scholar]
- Szajner, P.; Weisberg, A.S.; Moss, B. Physical and functional interactions between vaccinia virus F10 protein kinase and virion assembly proteins A30 and G7. J. Virol. 2004, 78, 266–274. [Google Scholar]
- Chiu, W.L.; Chang, W. Vaccinia virus J1R protein: A viral membrane protein that is essential for virion morphogenesis. J. Virol. 2002, 76, 9575–9587. [Google Scholar]
- Chiu, W.L.; Szajner, P.; Moss, B.; Chang, W. Effects of a temperature sensitivity mutation in the J1R protein component of a complex required for vaccinia virus assembly. J. Virol. 2005, 79, 8046–8056. [Google Scholar]
- Resch, W.; Weisberg, A.S.; Moss, B. Expression of the highly conserved vaccinia virus E6 protein is required for virion morphogenesis. Virology 2009, 386, 478–485. [Google Scholar]
- Boyd, O.; Turner, P.C.; Moyer, R.W.; Condit, R.C.; Moussatche, N. The E6 protein from vaccinia virus is required for the formation of immature virions. Virology 2010, 399, 201–211. [Google Scholar]
- Moss, B. Poxvirus DNA replication. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef]
- Garcia, A.D.; Moss, B. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J. Virol. 2001, 75, 6460–6471. [Google Scholar]
- Mutsafi, Y.; Fridmann-Sirkis, Y.; Milrot, E.; Hevroni, L.; Minsky, A. Infection cycles of large DNA viruses: Emerging themes and underlying questions. Virology 2014. [Google Scholar] [CrossRef]
- Ansarah-Sobrinho, C.; Moss, B. Role of the I7 protein in proteolytic processing of vaccinia virus membrane and core components. J. Virol. 2004, 78, 6335–6343. [Google Scholar]
- Byrd, C.M.; Hruby, D.E. A conditional-lethal vaccinia virus mutant demonstrates that the I7L gene product is required for virion morphogenesis. Virol. J. 2005, 2. [Google Scholar] [CrossRef]
- Satheshkumar, P.S.; Weisberg, A.S.; Moss, B. Vaccinia virus A19 protein participates in the transformation of spherical immature particles to barrel-shaped infectious virions. J. Virol. 2013, 87, 10700–10709. [Google Scholar]
- Howard, A.R.; Senkevich, T.G.; Moss, B. Vaccinia virus A26 and A27 proteins form a stable complex tethered to mature virions by association with the A17 transmembrane protein. J. Virol. 2008, 82, 12384–12391. [Google Scholar]
- Chichon, F.J.; Rodriguez, M.J.; Risco, C.; Fraile-Ramos, A.; Fernandez, J.J.; Esteban, M.; Carrascosa, J.L. Membrane remodelling during vaccinia virus morphogenesis. Biol. Cell 2009, 101, 401–414. [Google Scholar]
- Kuznetsov, Y.; Gershon, P.D.; McPherson, A. Atomic force microscopy investigation of vaccinia virus structure. J. Virol. 2008, 82, 7551–7566. [Google Scholar]
- Kato, S.E.M.; Condit, R.C.; Moussatche, N. The vaccinia virus E8R gene product is required for formation of transcriptionally active virions. Virology 2007, 367, 398–412. [Google Scholar]
- Kane, E.M.; Shuman, S. Temperature-sensitive mutations in the vaccinia virus-h4 gene encoding a component of the virion rna-polymerase. J. Virol. 1992, 66, 5752–5762. [Google Scholar]
- Zhang, Y.F.; Ahn, B.Y.; Moss, B. Targeting of a multicomponent transcription apparatus into assembling vaccinia virus-particles requires rap94, an rna polymerase-associated protein. J. Virol. 1994, 68, 1360–1370. [Google Scholar]
- McFadden, B.D.; Moussatche, N.; Kelley, K.; Kang, B.H.; Condit, R.C. Vaccinia virions deficient in transcription enzymes lack a nucleocapsid. Virology 2012, 434, 50–8. [Google Scholar]
- Byrd, C.M.; Hruby, D.E. Vaccinia virus proteolysis—A review. Rev. Med. Virol. 2006, 16, 187–202. [Google Scholar]
- Byrd, C.M.; Bolken, T.C.; Hruby, D.E. The vaccinia virus I7L gene product is the core protein proteinase. J. Virol. 2002, 76, 8973–8976. [Google Scholar]
- Kane, E.M.; Shuman, S. Vaccinia virus morphogenesis is blocked by a temperature-sensitive mutation in the I7 gene that encodes a virion component. J. Virol. 1993, 67, 2689–2698. [Google Scholar]
- Ansarah-Sobrinho, C.; Moss, B. Vaccinia virus G1 protein, a predicted metalloprotease, is essential for morphogenesis of infectious virions but not for cleavage of major core proteins. J. Virol. 2004, 78, 6855–6863. [Google Scholar]
- Hedengren-Olcott, M.; Byrd, C.M.; Watson, J.; Hruby, D.E. The vaccinia virus G1L putative metalloproteinase is essential for viral replication in vivo. J. Virol. 2004, 78, 9947–9953. [Google Scholar]
- Honeychurch, K.M.; Byrd, C.M.; Hruby, D.E. Mutational analysis of the potential catalytic residues of the VV GIL metalloproteinase. Virol. J. 2006, 3. [Google Scholar] [CrossRef]
- Alzhanova, D.; Hruby, D.E. A host cell membrane protein, golgin-97, is essential for poxvirus morphogenesis. Virology 2007, 362, 421–427. [Google Scholar]
- Alzhanova, D.; Hruby, D.E. A trans-golgi network resident protein, golgin-97, accumulates in viral factories and incorporates into virions during poxvirus infection. J. Virol. 2006, 80, 11520–11527. [Google Scholar]
- McNulty, S.; Bornmann, W.; Schriewer, J.; Werner, C.; Smith, S.K.; Olson, V.A.; Damon, I.K.; Buller, R.M.; Heuser, J.; Kalman, D. Multiple phosphatidylinositol 3-kinases regulate vaccinia virus morphogenesis. PLoS One 2010, 5, e10884. [Google Scholar]
- Senkevich, T.G.; Weisberg, A.S.; Moss, B. Vaccinia virus E10R protein is associated with the membranes of intracellular mature virions and has a role in morphogenesis. Virology 2000, 278, 244–252. [Google Scholar]
- Senkevich, T.G.; White, C.L.; Weisberg, A.; Granek, J.A.; Wolffe, E.J.; Koonin, E.V.; Moss, B. Expression of the vaccinia virus A2.5L redox protein is required for virion morphogenesis. Virology 2002, 300, 296–303. [Google Scholar]
- White, C.L.; Senkevich, T.G.; Moss, B. Vaccinia virus G4L glutaredoxin is an essential intermediate of a cytoplasmic disulfide bond pathway required for virion assembly. J. Virol. 2002, 76, 467–472. [Google Scholar]
- Hakim, M.; Fass, D. Cytosolic Disulfide Bond Formation in Cells Infected with Large Nucleocytoplasmic DNA Viruses. Antioxid. Redox Signal. 2010, 13, 1261–1271. [Google Scholar]
- Schmidt, F.I.; Bleck, C.K. E.; Reh, L.; Novy, K.; Wollscheid, B.; Helenius, A.; Stahlberg, H.; Mercer, J. Vaccinia Virus Entry Is Followed by Core Activation and Proteasome-Mediated Release of the Immunomodulatory Effector VH1 from Lateral Bodies. Cell Rep. 2013, 4, 464–476. [Google Scholar]
- Wickramasekera, N.T.; Traktman, P. Structure/Function analysis of the vaccinia virus F18 phosphoprotein, an abundant core component required for virion maturation and infectivity. J. Virol. 2010, 84, 6846–6860. [Google Scholar]
- Zhang, Y.F.; Moss, B. Vaccinia virus morphogenesis is interrupted when expression of the gene encoding an 11-kilodalton phosphorylated protein is prevented by the escherichia-coli lac repressor. J. Virol. 1991, 65, 6101–6110. [Google Scholar]
- Chen, Y.; Honeychurch, K.M.; Yang, G.; Byrd, C.M.; Harver, C.; Hruby, D.E.; Jordan, R. Vaccinia virus p37 interacts with host proteins associated with LE-derived transport vesicle biogenesis. Virol. J. 2009, 6. [Google Scholar] [CrossRef]
- Meseda, C.A.; Campbell, J.; Kumar, A.; Garcia, A.D.; Merchlinsky, M.; Weir, J.P. Effect of the deletion of genes encoding proteins of the extracellular virion form of vaccinia virus on vaccine immunogenicity and protective effectiveness in the mouse model. PLoS One 2013, 8, e67984. [Google Scholar]
- Horsington, J.; Lynn, H.; Turnbull, L.; Cheng, D.; Braet, F.; Diefenbach, R.J.; Whitchurch, C.B.; Karupiah, G.; Newsome, T.P. A36-dependent Actin Filament Nucleation Promotes Release of Vaccinia Virus. PLoS Pathog. 2013, 9, e1003239. [Google Scholar]
- Alvarez, D.E.; Agaisse, H. The formin FHOD1 and the small GTPase Rac1 promote vaccinia virus actin-based motility. J. Cell Biol. 2013, 202, 1075–1090. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Cooper, T.; Howley, P.M.; Hayball, J.D. From Crescent to Mature Virion: Vaccinia Virus Assembly and Maturation. Viruses 2014, 6, 3787-3808. https://doi.org/10.3390/v6103787
Liu L, Cooper T, Howley PM, Hayball JD. From Crescent to Mature Virion: Vaccinia Virus Assembly and Maturation. Viruses. 2014; 6(10):3787-3808. https://doi.org/10.3390/v6103787
Chicago/Turabian StyleLiu, Liang, Tamara Cooper, Paul M. Howley, and John D. Hayball. 2014. "From Crescent to Mature Virion: Vaccinia Virus Assembly and Maturation" Viruses 6, no. 10: 3787-3808. https://doi.org/10.3390/v6103787